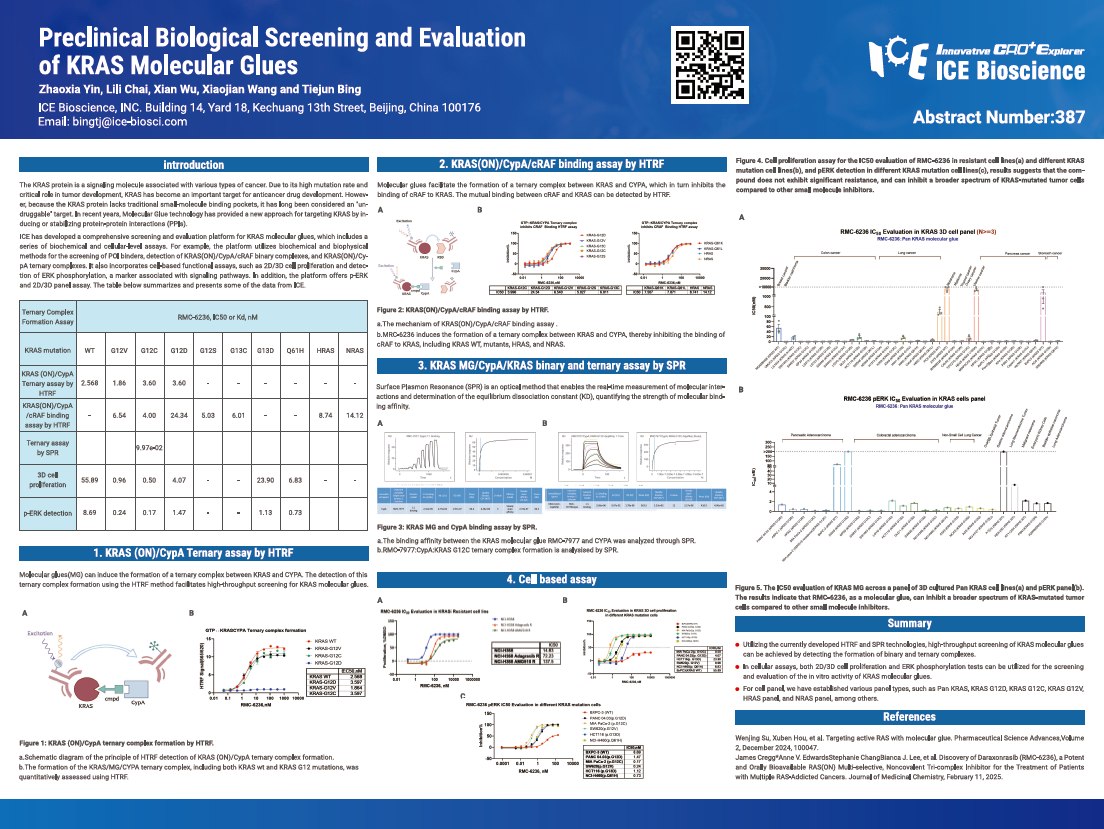
The KRAS protein is a signaling molecule associated with various types of cancer. Due to its high mutation rate and critical role in tumor development, KRAS has become an important target for anticancer drug development. However, because the KRAS protein lacks traditional small-molecule binding pockets, it has long been considered an "undruggable" target. In recent years, Molecular Glue technology has provided a new approach for targeting KRAS by inducing or stabilizing protein-protein interactions (PPls).
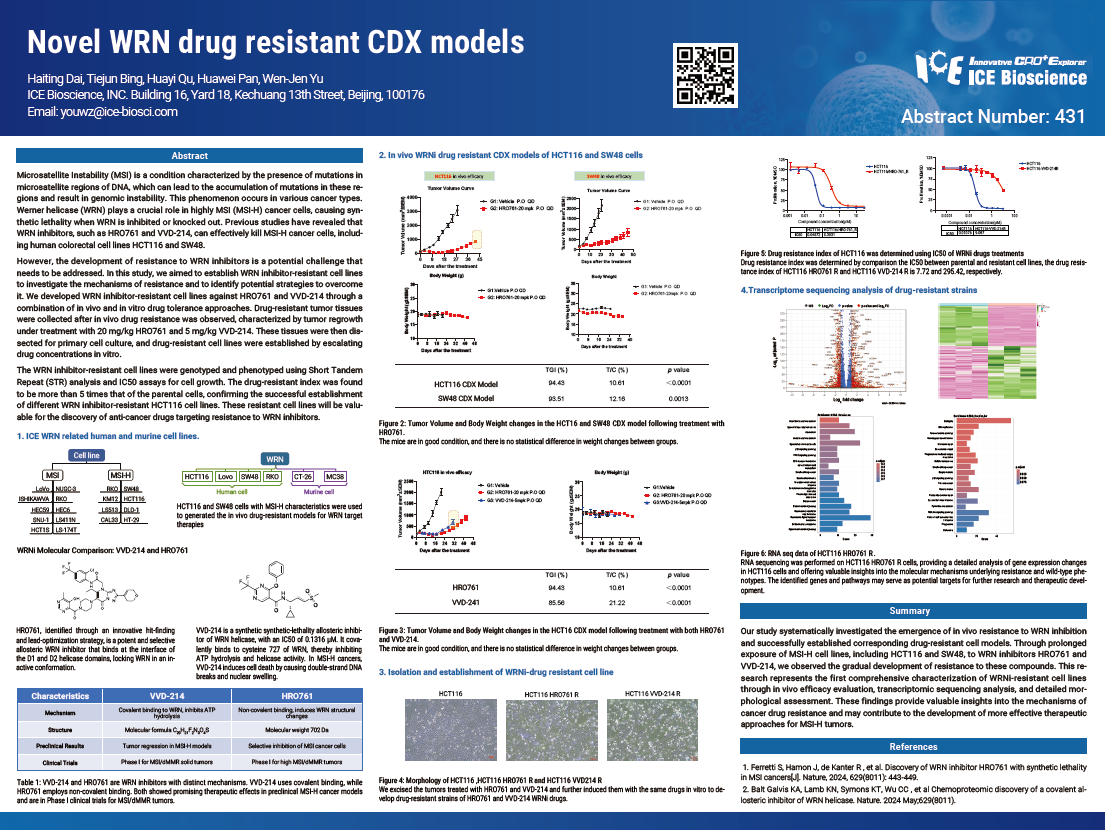
Microsatellite Instability (MSI) is a condition characterized by the presence of mutations in microsatellite regions of DNA, which can lead to the accumulation of mutations in these re- gions and result in genomic instability. This phenomenon occurs in various cancer types. Werner helicase (WRN) plays a crucial role in highly MSI (MSI-H) cancer cells, causing syn- thetic lethality when WRN is inhibited or knocked out. Previous studies have revealed that WRN inhibitors, such as HRO761 and VVD-214, can effectively kill MSI-H cancer cells, includ- ing human colorectal cell lines HCT116 and SW48.
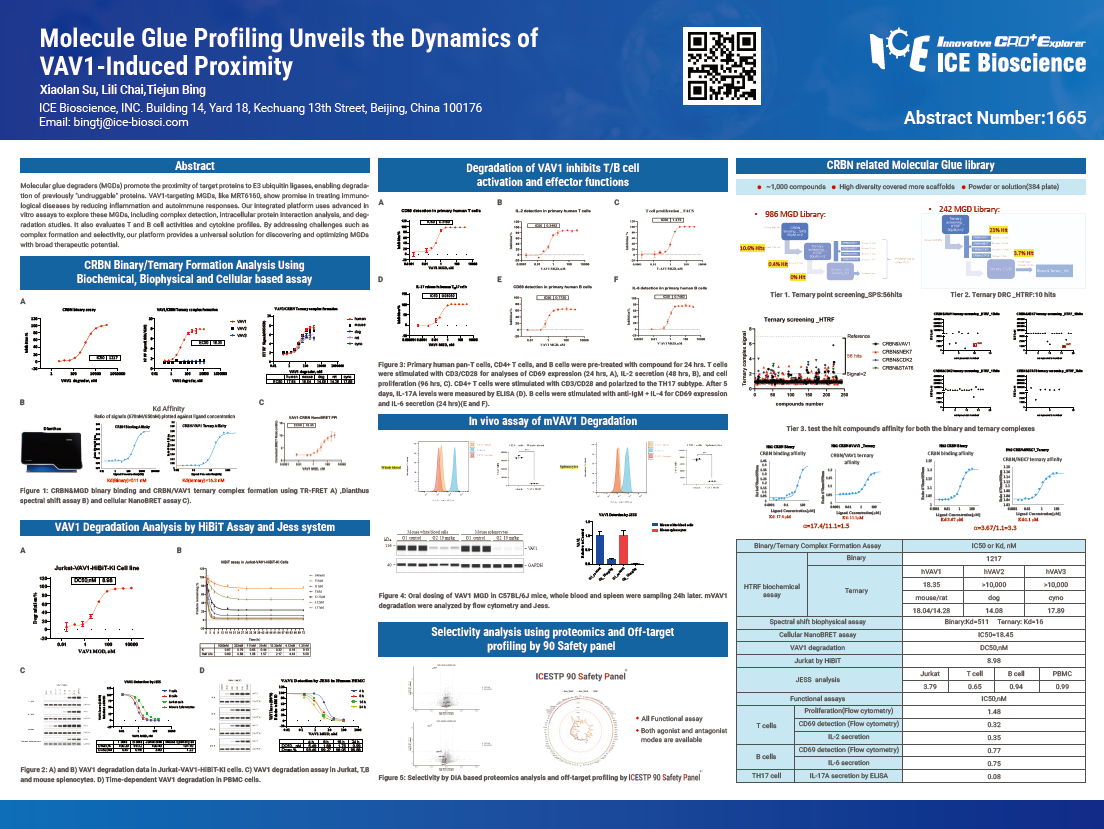
Molecular glue degraders (MGDs) promote the proximity of target proteins to E3 ubiquitin ligases, enabling degradation of previously "undruggable" proteins. VAV1-targeting MGDs, like MRT6160, show promise in treating immunological diseases by reducing inflammation and autoimmune responses. Our integrated platform uses advanced in vitro assays to explore these MGDs, including complex detection, intracellular protein interaction analysis, and degradation studies. It also evaluates T and B cell activities and cytokine profiles. By addressing challenges such as complex formation and selectivity, our platform provides a universal solution for discovering and optimizing MGDs with broad therapeutic potential.
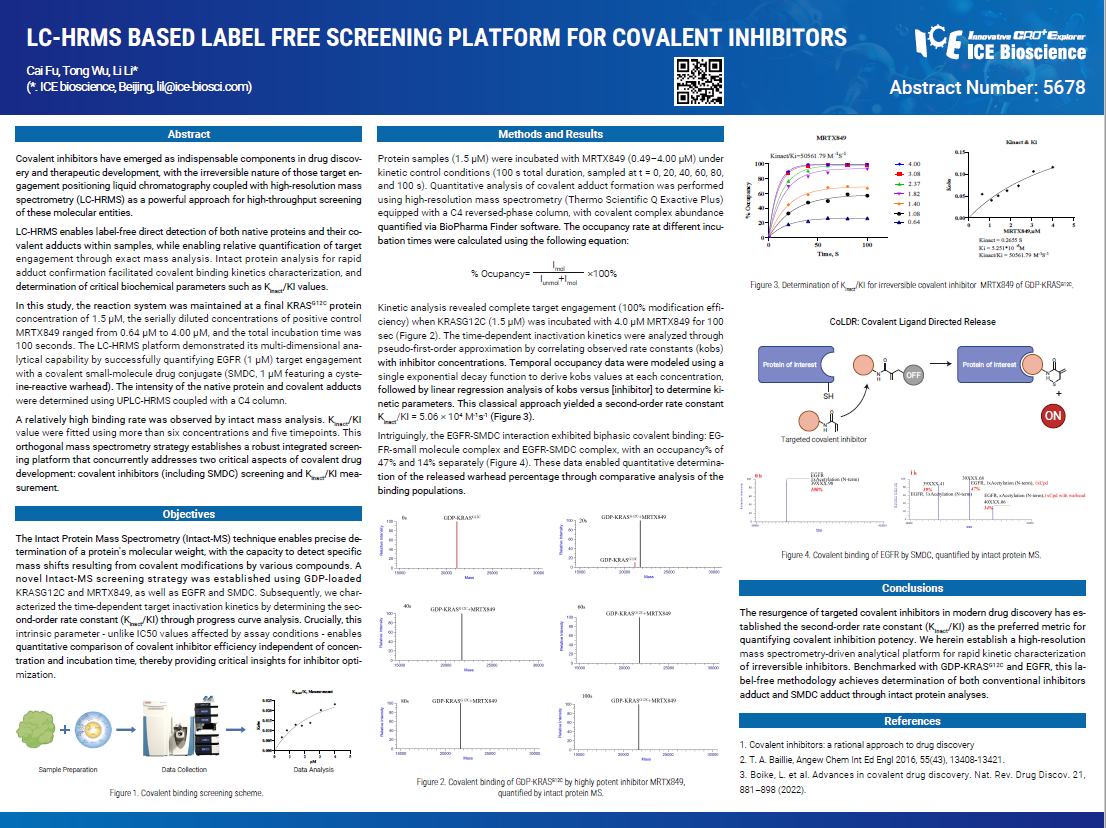
Covalent inhibitors have emerged as indispensable components in drug discov- ery and therapeutic development, with the irreversible nature of those target en- gagement positioning liquid chromatography coupled with high-resolution mass spectrometry (LC-HRMS) as a powerful approach for high-throughput screening of these molecular entities.
Address: Bldg 16, Yd 18, Kechuang 13th St, Etown, Tongzhou Dist, Beijing, 100176, China
Email: marketing@ice-biosci.com
Tel:+86-10-67809840
