The article was co-published by Hainan University, ICE Bioscience and so on.
Abstract
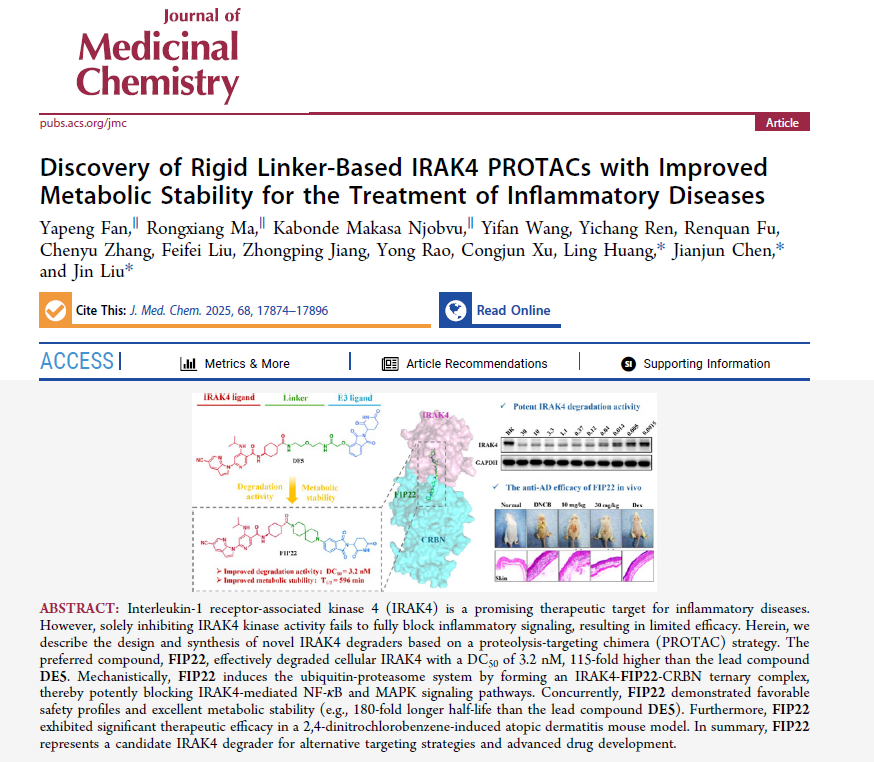
Interleukin-1 receptor-associated kinase 4 (IRAK4) is a promising therapeutic target for inflammatory diseases. However, solely inhibiting IRAK4 kinase activity fails to fully block inflammatory signaling, resulting in limited efficacy. Herein, we describe the design and synthesis of novel IRAK4 degraders based on a proteolysis-targeting chimera (PROTAC) strategy. The preferred compound, FIP22, effectively degraded cellular IRAK4 with a DC50 of 3.2 nM, 115-fold higher than the lead compound DE5. Mechanistically, FIP22 induces the ubiquitin-proteasome system by forming an IRAK4-FIP22-CRBN ternary complex, thereby potently blocking IRAK4-mediated NF-κB and MAPK signaling pathways. Concurrently, FIP22 demonstrated favorable safety profiles and excellent metabolic stability (e.g., 180-fold longer half-life than the lead compound DE5). Furthermore, FIP22 exhibited significant therapeutic efficacy in a 2,4-dinitrochlorobenzene-induced atopic dermatitis mouse model. In summary, FIP22 represents a candidate IRAK4 degrader for alternative targeting strategies and advanced drug development.
The article was co-published by China Pharmaceutical University and ICE Bioscience.
Abstract
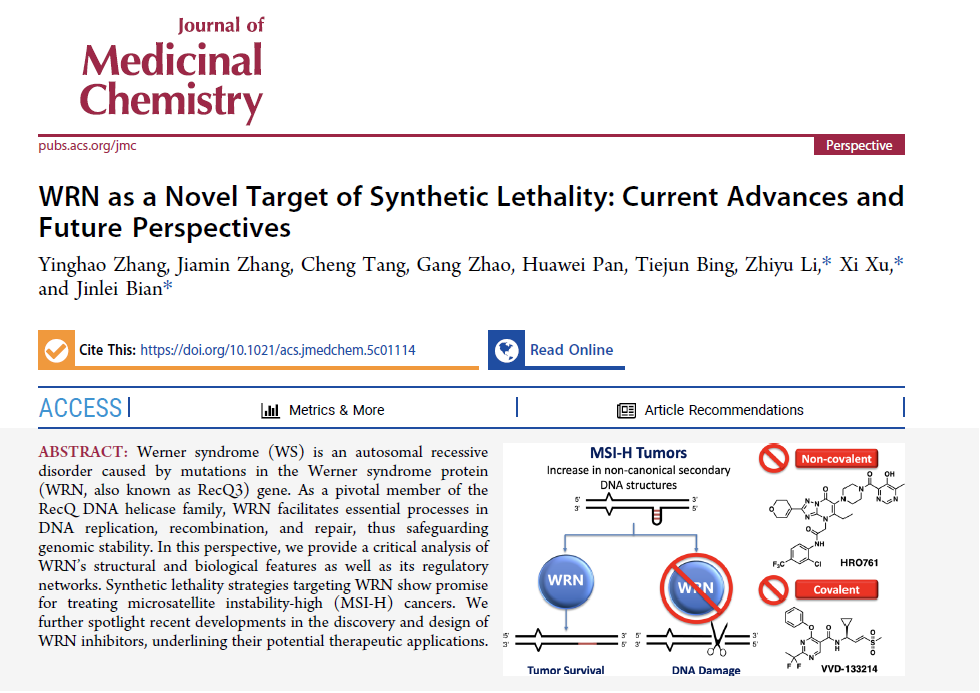
Werner syndrome (WS) is an autosomal recessive disorder caused by mutations in the Werner syndrome protein (WRN, also known as RecQ3) gene. As a pivotal member of the RecQ DNA helicase family, WRN facilitates essential processes in DNA replication, recombination, and repair, thus safeguarding genomic stability. In this perspective, we provide a critical analysis of WRN’s structural and biological features as well as its regulatory networks. Synthetic lethality strategies targeting WRN show promise for treating microsatellite instability-high (MSI-H) cancers. We further spotlight recent developments in the discovery and design of WRN inhibitors, underlining their potential therapeutic applications.
The article was co-published by China Pharmaceutical University and ICE Bioscience.
Abstract
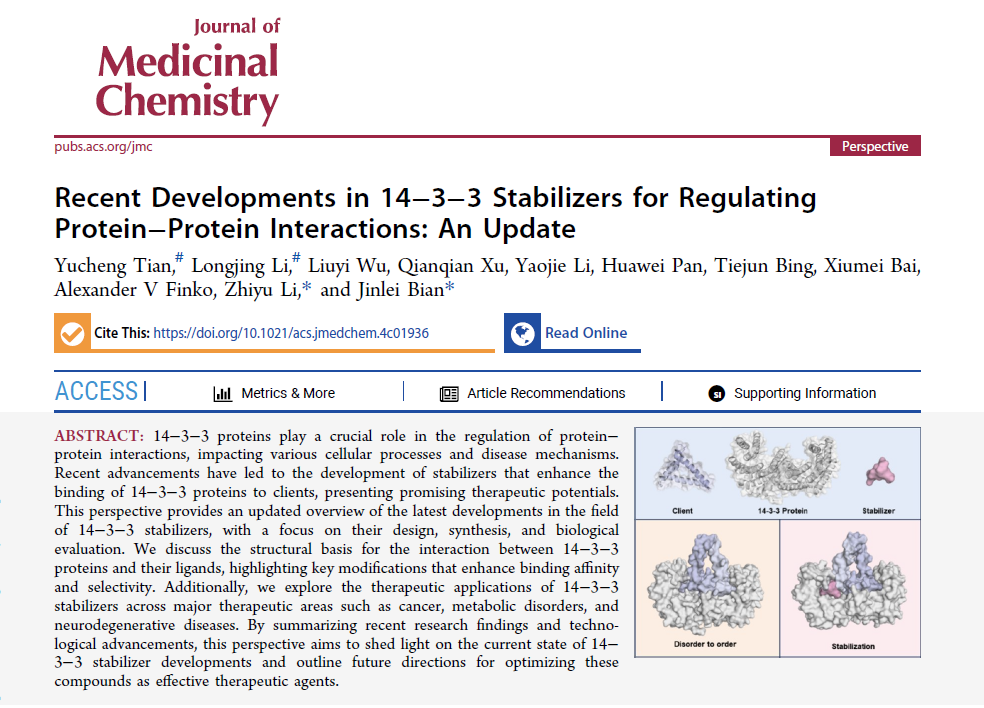
14−3−3 proteins play a crucial role in the regulation of protein− protein interactions, impacting various cellular processes and disease mechanisms. Recent advancements have led to the development of stabilizers that enhance the binding of 14−3−3 proteins to clients, presenting promising therapeutic potentials. This perspective provides an updated overview of the latest developments in the field of 14−3−3 stabilizers, with a focus on their design, synthesis, and biological evaluation. We discuss the structural basis for the interaction between 14−3−3 proteins and their ligands, highlighting key modifications that enhance binding affinity and selectivity. Additionally, we explore the therapeutic applications of 14−3−3 stabilizers across major therapeutic areas such as cancer, metabolic disorders, and neurodegenerative diseases. By summarizing recent research findings and technological advancements, this perspective aims to shed light on the current state of 14−3−3 stabilizer developments and outline future directions for optimizing these compounds as effective therapeutic agents.

The article was co-published by Beijing University of Chemical Technology and ICE Bioscience.
Abstract
Alzheimer’s disease is the predominant form of dementia, and disulfidptosis is the latest reported mode of cell death that impacts various disease processes. This study used bioinformatics to analyze genes associated with disulfidptosis in Alzheimer’s disease comprehensively. Based on the public datasets, the differentially expressed genes associated with disulfidptosis were identified, and immune cell infiltration was investigated through correlation analysis. Subsequently, hub genes were determined by a randomforest model. A prediction model was constructed using logistic regression. In addition, the drug-target affinity was predicted by a graph neural network model, and the results were validated by molecular docking. Five hub genes (PPEF1, NEUROD6, VIP, NUPR1, and GEM) were identified. The gene set showed significant enrichment for AD-related pathways. The logistic regression model demonstrated an AUC of 0.952, with AUC values of 0.916 and 0.864 in validated datasets. The immune infiltration analysis revealed significant heterogeneity between the Alzheimer’s disease and control groups. High-affinity drugs for hub genes were identified. Through our study, a disease prediction model was constructed using potential biomarkers, and drugs targeting the genes were predicted. These results contribute to further understanding of the molecular mechanisms underlying Alzheimer’s disease.