Signal Transducer and Activator of Transcription 6 (STAT6) is a pivotal transcription factor activated downstream of the interleukin-4 (IL-4) and interleukin-13 (IL-13) signaling pathways. It governs the expression of key genes involved in type 2 immune responses, including Th2 cell differentiation, IgE class switching, and M2 macrophage polarization. Dysregulated STAT6 activity has been implicated in a range of diseases, from asthma and atopic dermatitis to fibrosis and immune-evasive tumors.
With the clinical advancement of STAT6-targeting drugs like Kymera's KT-621 and Recludix's REX-8756, the demand for precise STAT6 binding profiling is rising—supporting target validation, mechanism studies, and therapeutic development.
We developed TR-FRET assays to evaluate compound binding to STAT6. For degrader molecules, we also performed binary and ternary complex formation assays to quantify target–ligase engagement and cooperativity.
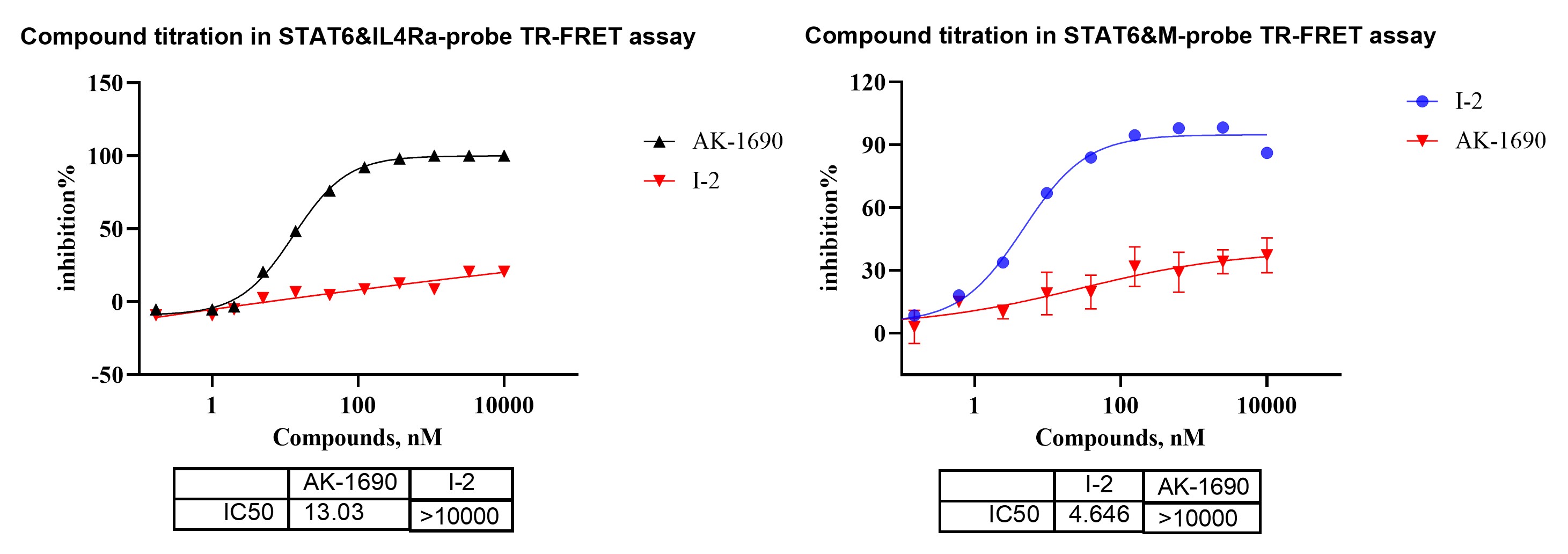
Figure 1.HTRF STAT6 probe-displacement assay using IL-4Rα and M-probe formats to profile orthosteric and allosteric STAT6 binders.
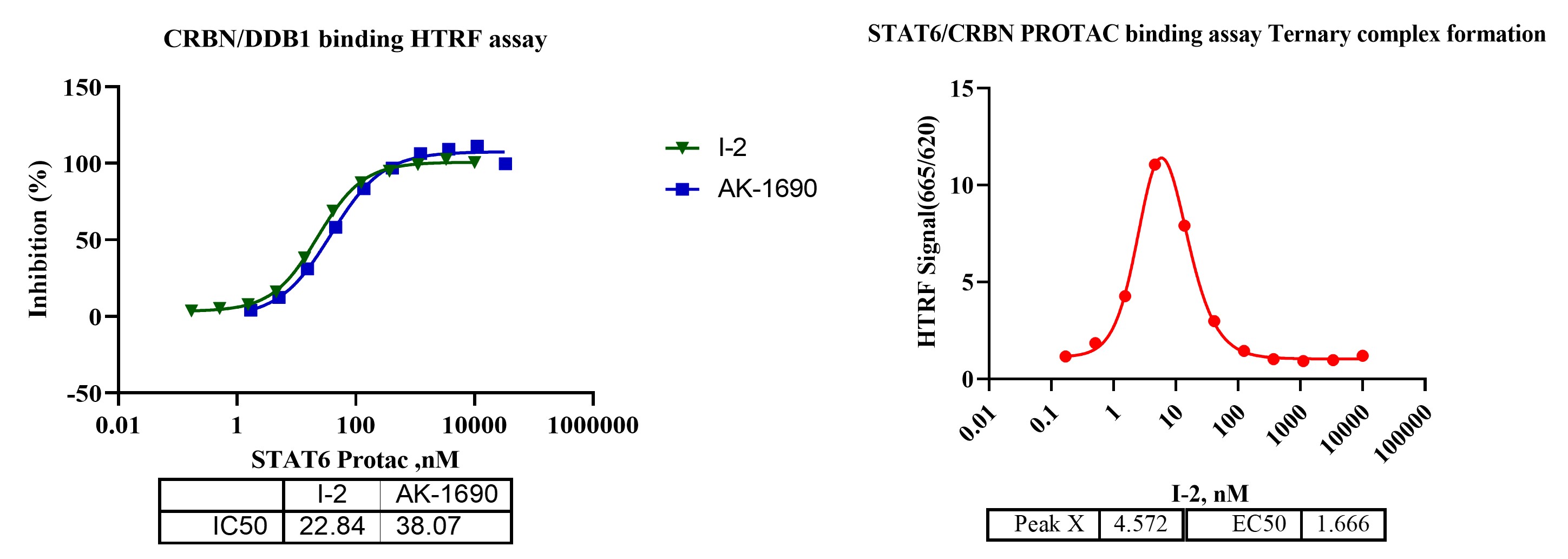
Figure 2. TR-FRET-based binary and ternary complex formation for STAT6 degraders.
To accurately characterize binding kinetics and selectivity of STAT6-targeting compounds, we employ both Surface Plasmon Resonance (SPR) and spectral shift–based assays. This dual-platform strategy allows us to balance sensitivity, throughput, and molecular mechanism insights, especially when dealing with weak-to-moderate SH2-domain interactions and structurally diverse chemotypes such as degraders or allosteric inhibitors.
To accurately characterize binding kinetics and selectivity of STAT6-targeting compounds, we employ both Surface Plasmon Resonance (SPR) and spectral shift–based assays. This dual-platform strategy allows us to balance sensitivity, throughput, and molecular mechanism insights, especially when dealing with weak-to-moderate SH2-domain interactions and structurally diverse chemotypes such as degraders or allosteric inhibitors.
For STAT6, SPR provides real-time, label-free kinetic profiling and is particularly well-suited for measuring binary binding affinity (KD) and association/dissociation rates (ka/kd). To support selectivity profiling, we conducted a STAT family SPR panel using compound SI-109, comparing its binding across STAT1–STAT6.
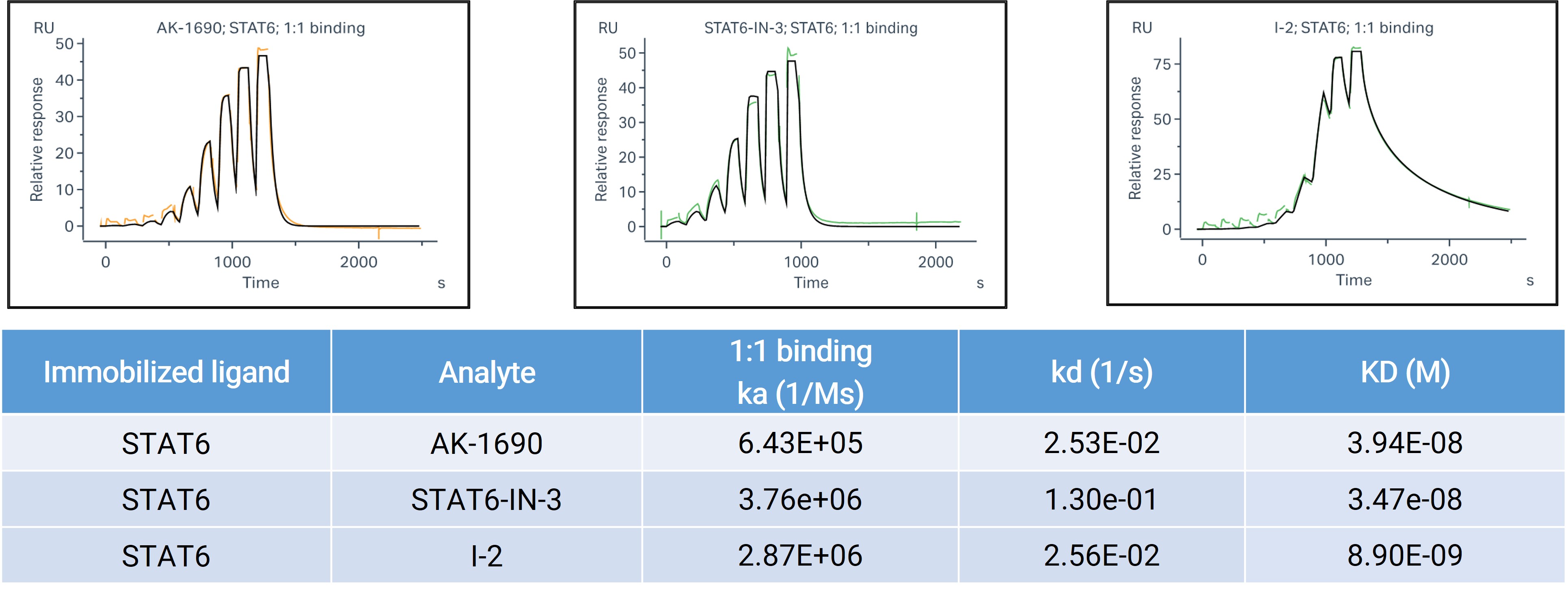
Figure 3. SPR binding analysis of STAT6.
SpS (spectral shift)-based binding assays quantify interaction between CRBN, PROTACs (AK-1690, I-2), and STAT6. Ternary complex formation significantly enhances binding affinity compared to binary CRBN–PROTAC binding, with a 99-fold cooperativity for AK-1690 and 9.2-fold for I-2. Assay performed in 384-well format.
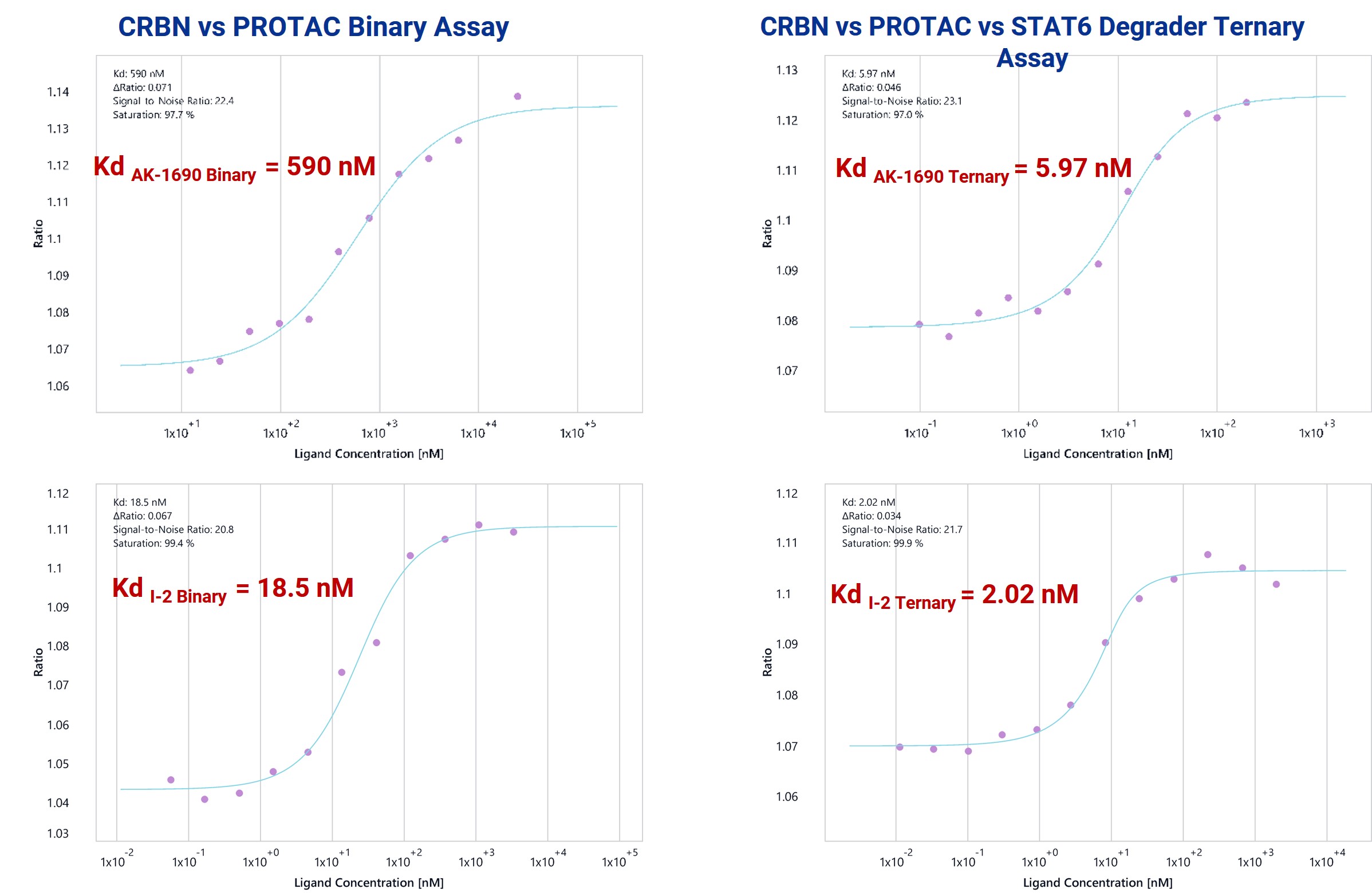
Figure 4. Sps-based binary and ternary complex formation assays for STAT6 degraders.
To confirm functional degradation of STAT6 inside cells, we employed a multi-assay validation strategy combining HiBiT-tag quantification, flow cytometry, and Western blotting. This integrated approach ensures high sensitivity, reproducibility, and orthogonal validation of degradation activity.
This is particularly critical for STAT6, where transcriptional feedback, protein turnover rate, and cell-type specificity can influence readouts. Multi-assay confirmation ensures observed degradation is real, robust, and mechanistically consistent.
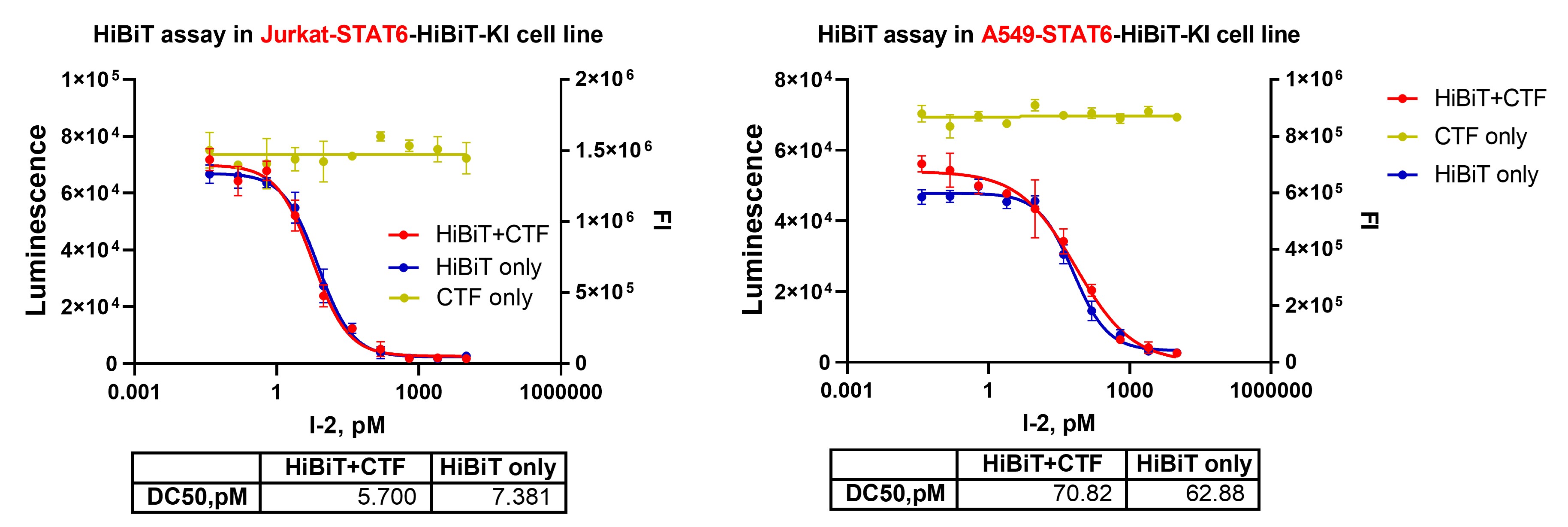
Figure 5. HiBiT-STAT6 degradation measured simultaneously with CellTiter-Fluor™–based viability. Luminescent HiBiT signal and fluorescence-based CellTiter-Fluor™ viability were acquired in the same assay to avoid luminescent signal interference.
STAT6 protein levels were detected by flow cytometry following treatment with degrader I-2 at increasing concentrations.
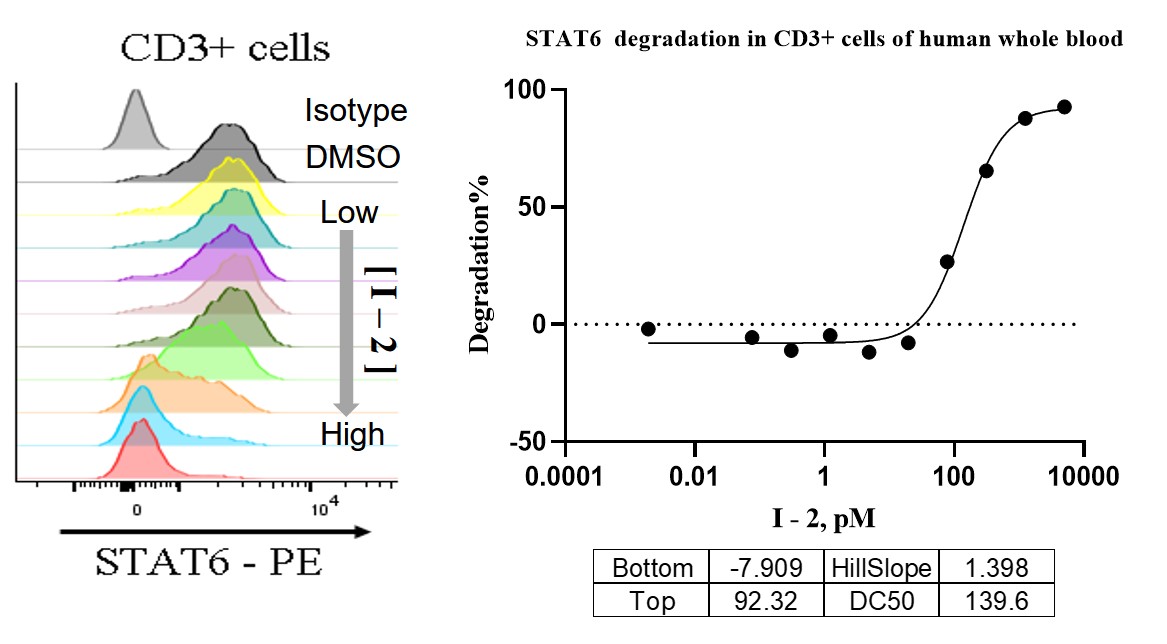
Figure 6. Flow cytometry analysis of STAT6 levels in CD3⁺ T cells.
To independently validate STAT6 degradation and assess endogenous protein levels across cell types, we performed Western blot analysis in both cell lines (Jurkat, A549) and primary cells (PBMCs). As a gold-standard method, Western blot provides direct visualization of target protein abundance, supporting the results from HiBiT and flow cytometry with orthogonal evidence.

Figure 7-1. Western blot confirmation of STAT6 degradation in multiple cell types.
Primary human smooth muscle cells provide a physiologically relevant context for evaluating STAT6 degraders. Unlike engineered reporter lines, these cells maintain native STAT6 expression levels, intact ubiquitin–proteasome machinery, and tissue-specific regulatory signaling, allowing a more accurate assessment of degrader activity in disease-relevant cell types.
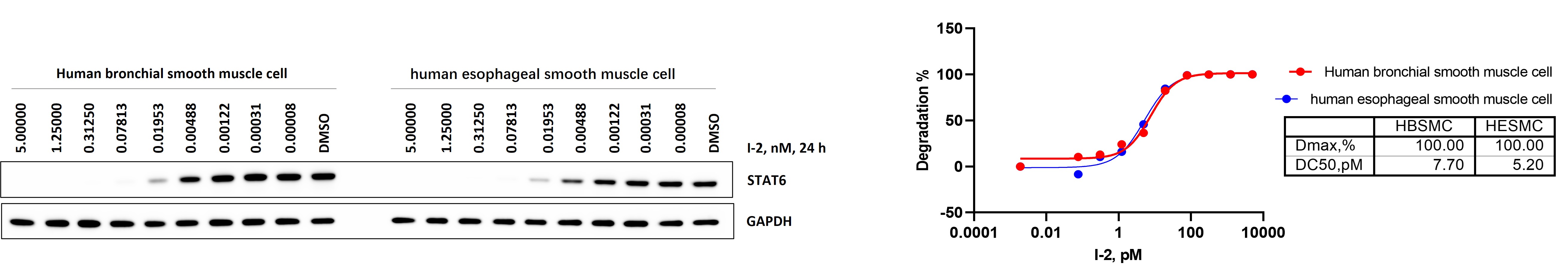
Figure 7-2. STAT6 degradation in primary human bronchial (HBSMC) and esophageal (HESMC) smooth muscle cells.
Demonstrating target degradation is only part of the story—verifying downstream functional impact is critical to validating STAT6 as a therapeutic target. As a key transcription factor in the IL-4/IL-13 signaling axis, STAT6 drives expression of multiple disease-relevant genes in immune cells, including CCL17 (TARC) and CD23, and regulates its activity through phosphorylation-dependent nuclear translocation.
Primary immune cells exhibit robust IL-4–driven STAT6 activation, making PBMCs and whole blood suitable ex vivo systems for evaluating pharmacological suppression of the pathway. Flow-cytometric quantification of pSTAT6 provides a sensitive, cell-level functional readout to assess compound activity in physiologically relevant immune populations.
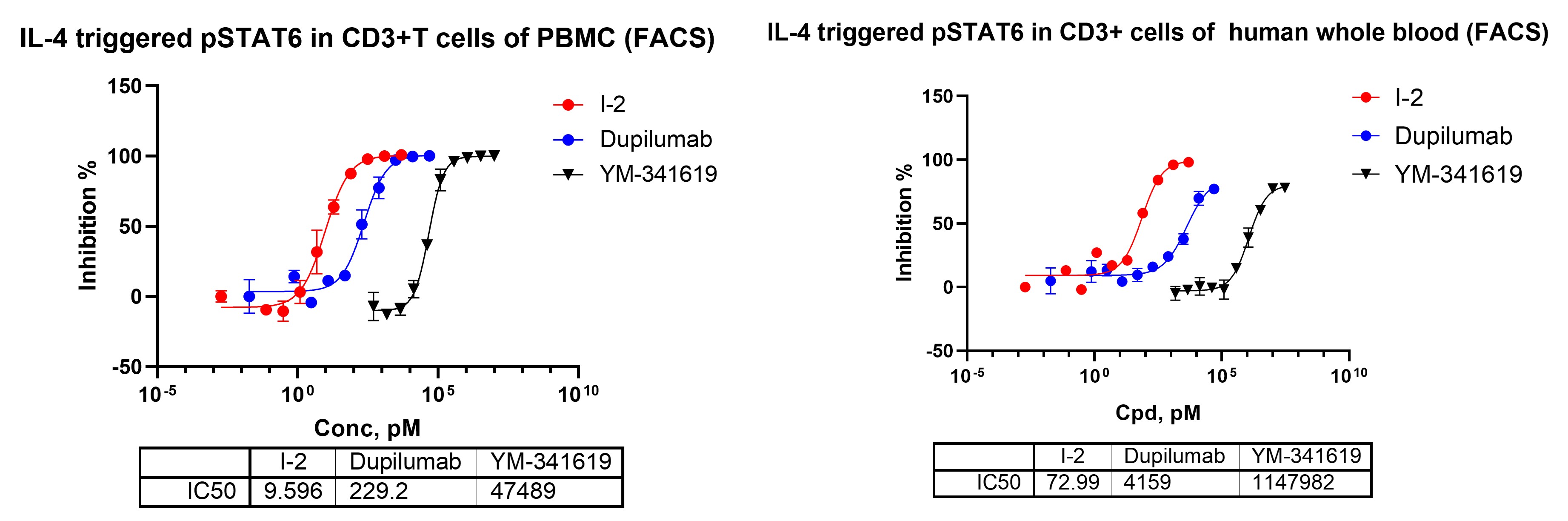
Figure 8. Flow-cytometric analysis of IL-4–induced STAT6 phosphorylation in CD3+ T cells of human PBMCs and whole blood.
Periostin is a downstream secreted biomarker strongly induced by IL-4 and IL-13 signaling in airway smooth muscle cells and is widely used as a functional readout of type-2 cytokine activation. Measuring periostin release in human bronchial smooth muscle cells provides a physiologically relevant assay to evaluate compound-mediated suppression of IL-4/IL-13–driven type-2 inflammatory responses.
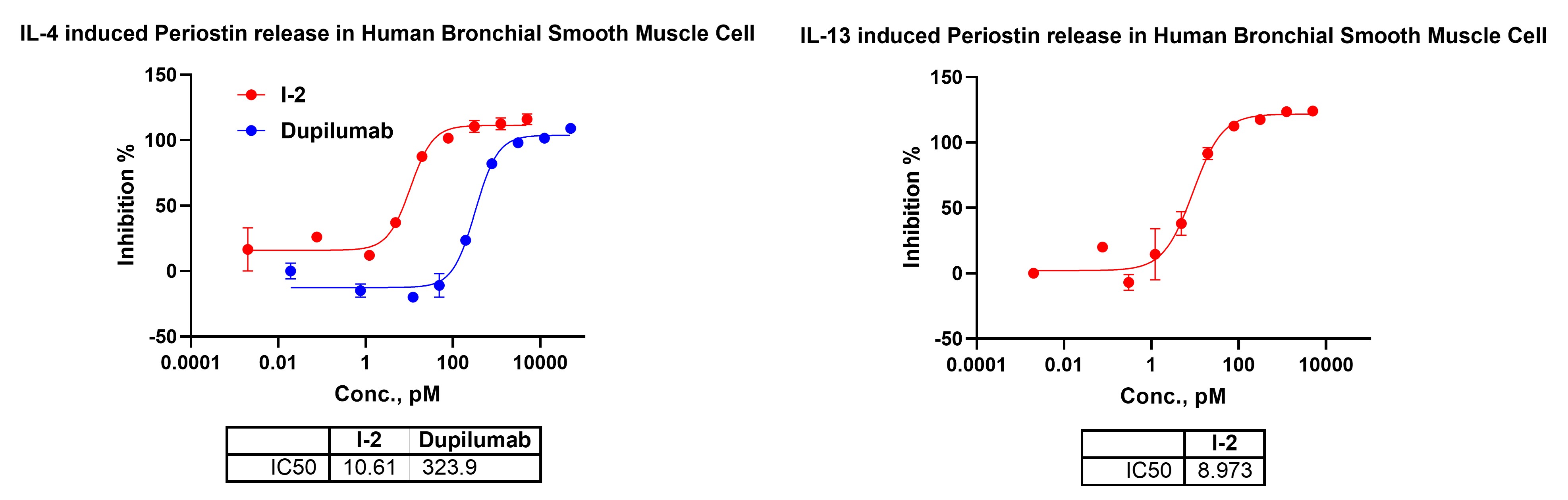
Figure 9. IL-4– and IL-13–stimulated periostin release in human bronchial smooth muscle cells.
IL-4 and IL-13 activate STAT6 across multiple immune pathways, driving distinct downstream responses such as reporter activation, chemokine release, and B-cell marker expression. Evaluating these diverse readouts enables a comprehensive assessment of how STAT6-targeting compounds modulate type-2 cytokine signaling across different cellular contexts.
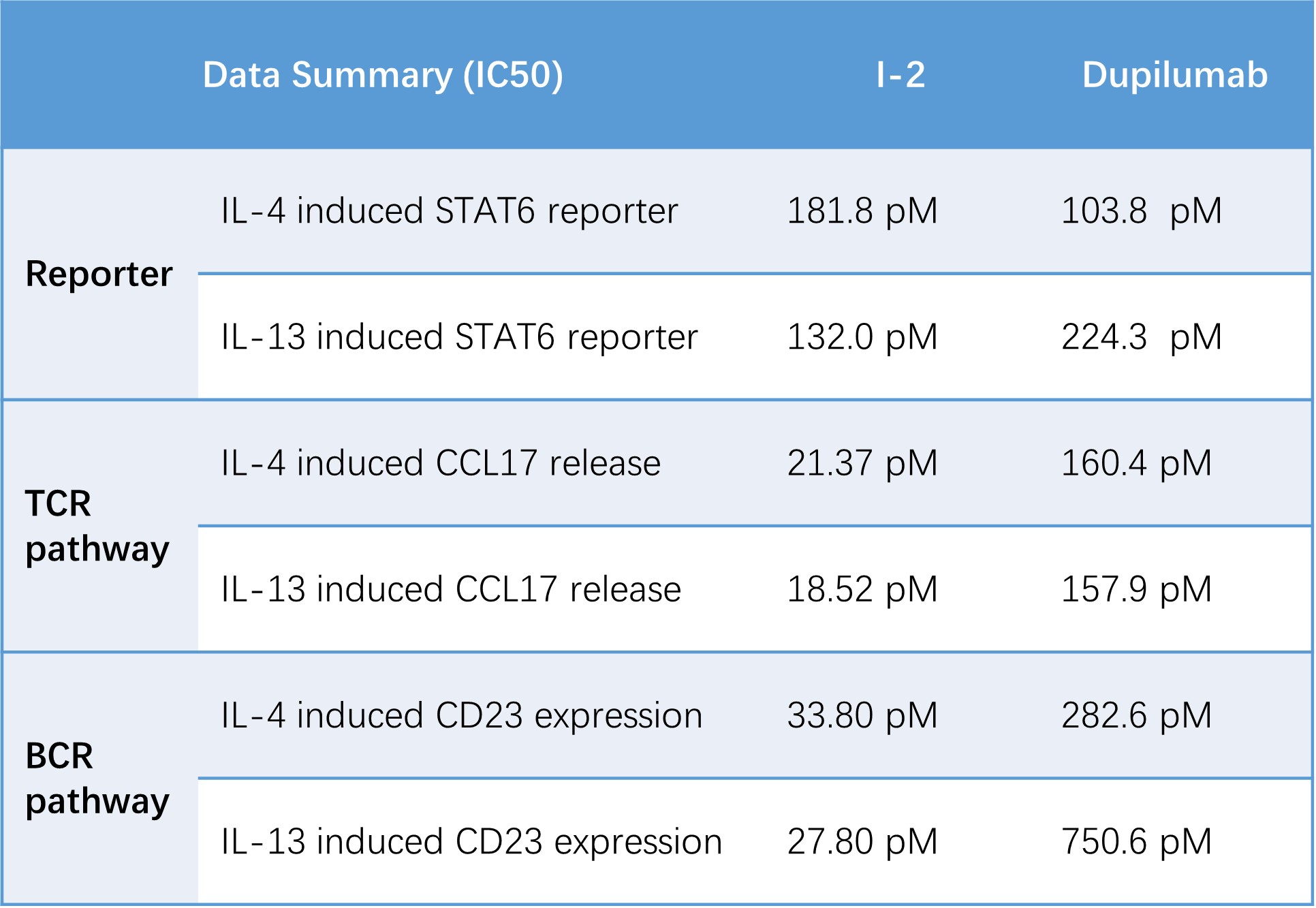
Table 1. Summary of STAT6-dependent functional assays across multiple cellular systems. Potency values (IC₅₀) were derived from three assay platforms: IL-4/IL-13-induced STAT6-Luc2P reporter activation in HEK293-STAT6-Luc2P cells, CCL17 secretion in IL-4/IL-13-stimulated human PBMCs, and CD23 upregulation in IL-4/IL-13-activated primary B cells.
Together, these biochemical, biophysical, and cellular assays establish a robust framework for characterizing STAT6-targeting molecules. By integrating binding, ternary complex formation, cellular degradation, and downstream functional readouts across multiple human cell systems, we provide a comprehensive yet practical platform for evaluating STAT6 degraders and inhibitors in IL-4/IL-13–driven biology.
2025-10-30
2025-10-23
2025-09-28
2025-08-19
We value your inquiries and are here to provide you with tailored solutions for your drug discovery and development needs. Whether you have questions, require more information, or are interested in discussing potential collaborations, our team of experts is just a message away.
Feel free to reach out to us.