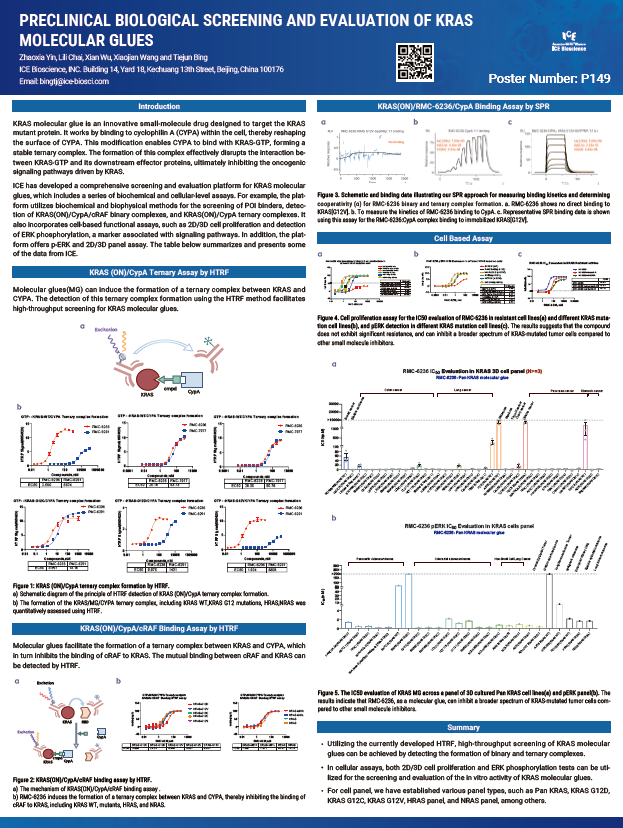
KRAS molecular glue is an innovative small-molecule drug designed to target the KRAS mutant protein. It works by binding to cyclophilin A (CYPA) within the cell, thereby reshaping the surface of CYPA. This modification enables CYPA to bind with KRAS-GTP, forming a stable ternary complex. The formation of this complex effectively disrupts the interaction be- tween KRAS-GTP and its downstream effector proteins, ultimately inhibiting the oncogenic signaling pathways driven by KRAS.
ICE has developed a comprehensive screening and evaluation platform for KRAS molecular glues, which includes a series of biochemical and cellular-level assays. For example, the plat- form utilizes biochemical and biophysical methods for the screening of POI binders, detec- tion of KRAS(ON)/CypA/cRAF binary complexes, and KRAS(ON)/CypA ternary complexes. It also incorporates cell-based functional assays, such as 2D/3D cell proliferation and detection of ERK phosphorylation, a marker associated with signaling pathways. In addition, the plat- form offers p-ERK and 2D/3D panel assay. The table below summarizes and presents some of the data from ICE.

Fragment-based drug design (FBDD) has emerged as a powerful strategy in drug discovery, particularly for identifying novel scaffolds and binding sites for challenging targets. We have employed a combination of Spectral Shift and TR-FRET technologies to discover new molecular glue scaffolds. Our fragment library is diverse and includes a rich variety of E3 ligases from different species, providing a sustainable and versatile screening platform.
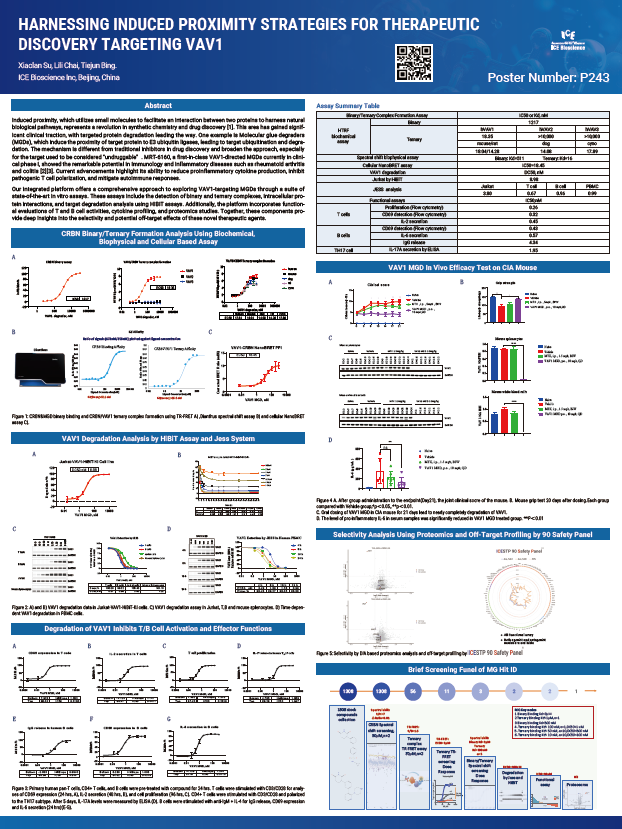
Induced proximity, which utilizes small molecules to facilitate an interaction between two proteins to harness natural biological pathways, represents a revolution in synthetic chemistry and drug discovery [1]. This area has gained significant clinical traction, with targeted protein degradation leading the way. One example is Molecular glue degraders (MGDs), which induce the proximity of target protein to E3 ubiquitin ligases, leading to target ubiquitination and degradation. The mechanism is different from traditional inhibitors in drug discovery and broaden the approach, especially for the target used to be considered "undruggable". MRT-6160, a first-in-class VAV1-directed MGDs currently in clinical phase I, showed the remarkable potential in Immunology and inflammatory diseases such as rheumatoid arthritis and colitis [2][3]. Current advancements highlight its ability to reduce proinflammatory cytokine production, inhibit pathogenic T cell polarization, and mitigate autoimmune responses.

In the development of new drugs, it is crucial to know the safety profile of drug candidates in advance, as this can prevent the drug from being withdrawn from the market due to safety concerns after launch, thus reducing the risk for pharmaceutical companies.
It is estimated that approximately 75% of all adverse drug reactions (ADRs) are dose-dependent type A reactions, which can be predicted according to the pharmacological profiles of drug candidates. The pharmacological profiles are mainly divided into primary effects, which are related to the action of the compound at its intended target, and secondary effects, which arise from interactions with non-primary targets, i.e. off-target effects. Off-target interactions are often the cause of ADRs in animal models or clinical studies, so careful characterization and identification of the secondary pharmacological profiles of drug candidates early in the drug discovery process could help reduce the incidence of type A ADRs.