Covalent drugs have emerged as a powerful class of therapeutics in targeted therapy, offering several significant advantages, including high selectivity due to their precise binding to specific amino acid residues, prolonged duration of action through irreversible binding, and reduced likelihood of drug resistance by maintaining efficacy despite genetic mutations.
A prime example of mass spectrometry in covalent binding analysis is identifying KRAS-G12C and AMG-510 conjugates. KRAS-G12C, a mutant form of the KRAS protein, is a key cancer therapy target due to its role in driving tumor growth. AMG-510 (sotorasib) is a pioneering covalent inhibitor designed to specifically bind to KRAS-G12C, irreversibly inhibiting its activity. Mass spectrometry accurately detects the covalent conjugates by identifying specific mass shifts indicating bond formation between AMG-510 and the cysteine residue on KRAS-G12C.
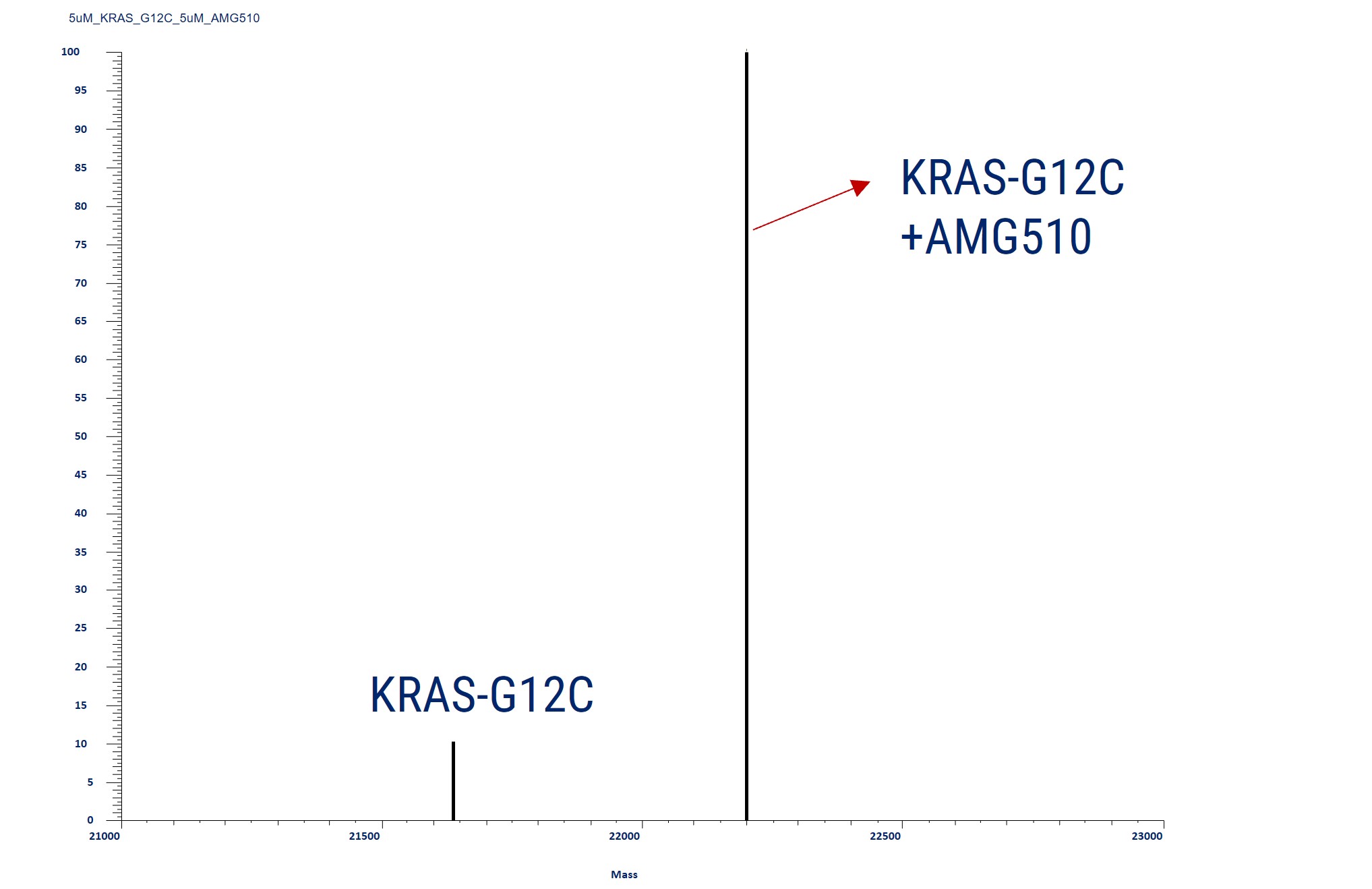
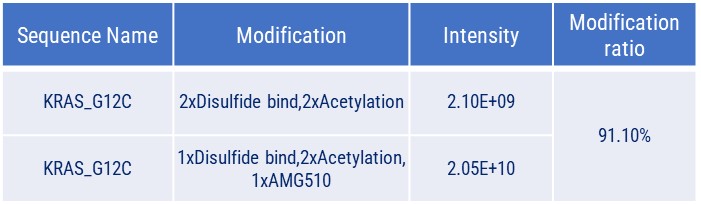
Covalent Binding Analysis of Acetylated KRAS-G12C. The mass spectrometry profiles reveal acetylated KRAS-G12C both before and after covalent modification with a compound. The graph shows the mass of the acetylated KRAS-G12C with and without the compound, indicating the acetylated KRAS-G12C with the AMG510 compound covalently attached.
We also provide covalent binding sample data using our expressed EGFR protein with Afatinib, WRN with reference compounds, and P53 and P53 Y220C with KG13. Contact us for detailed data and further information.
Our Kinact/Ki Assay Services provide comprehensive evaluation of covalent inhibitors, combining advanced mass spectrometry data analysis and model fitting to determine key kinetic parameters. This assay is crucial for understanding the reactivity and efficiency of covalent drugs.
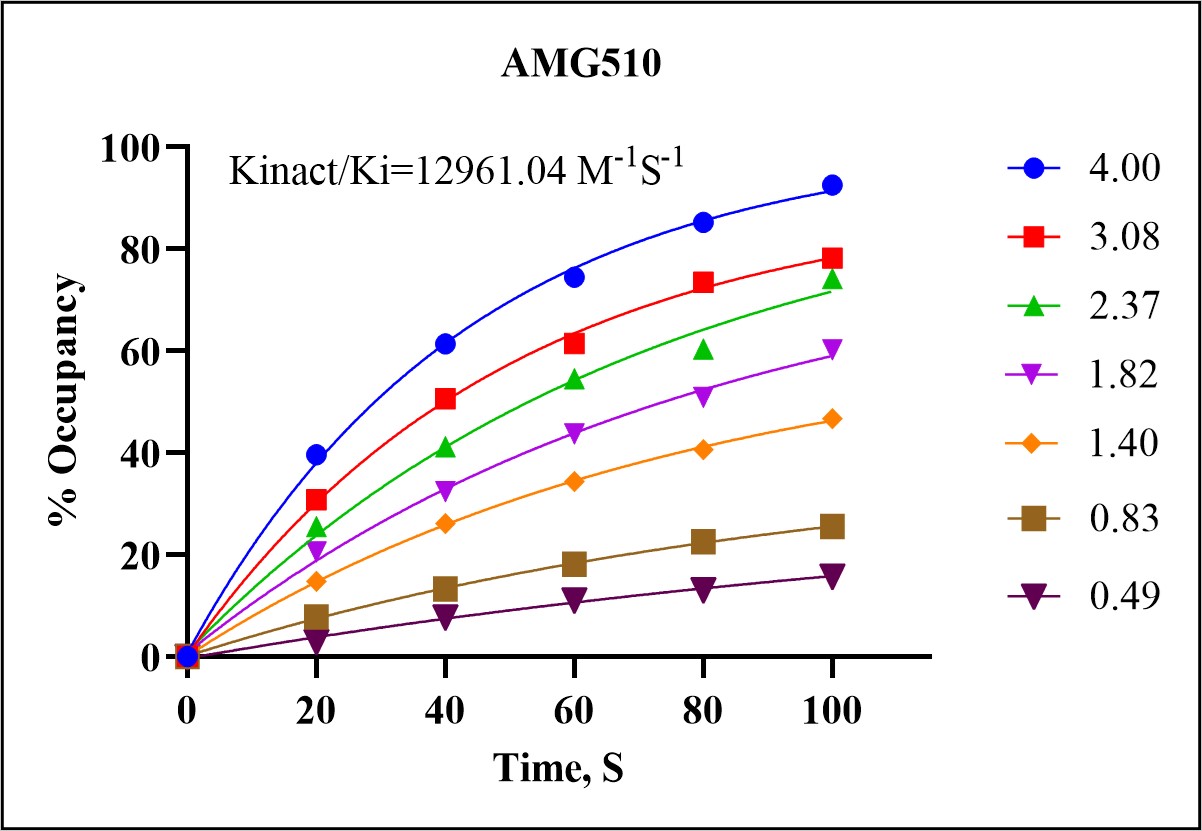
The Kinact/Ki assay measures the inactivation rate constant (Kinact) and the inhibition constant (Ki) for covalent inhibitors. These parameters are essential for characterizing the potency and binding kinetics of a drug, providing insights into its potential efficacy and selectivity.
In a demonstration of our Kinact/Ki assay capabilities, we analyzed the covalent interaction between the KRAS-G12C mutant protein and the covalent inhibitor AMG510 (sotorasib). The assay involved the following steps:
☑️ Incubation and Sampling
☑️ Mass Spectrometry Analysis
☑️ Data Analysis and Model Fitting
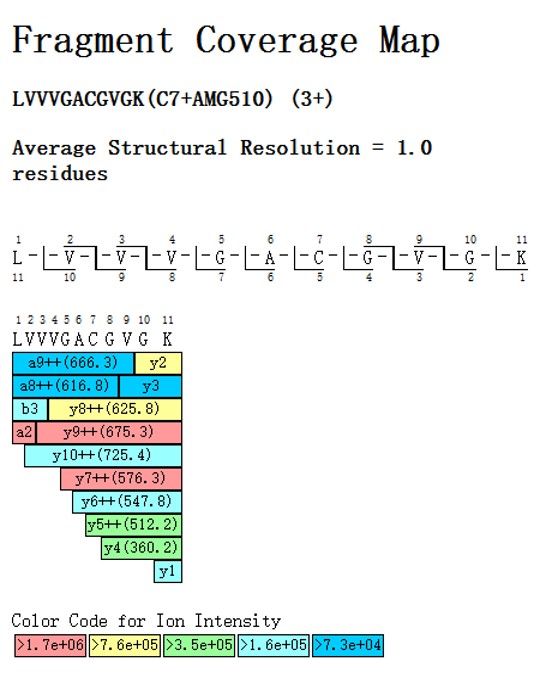
Our services provide detailed analysis of specific amino acid residues involved in covalent drug binding. This service is essential for understanding the precise interactions between covalent inhibitors and their target proteins, aiding in the optimization and development of effective therapeutics.
In a recent analysis of the KRASG12C mutant protein with the covalent inhibitor AMG510, our services identified the exact binding site of AMG510 on the KRASG12C protein. The process involved:
☑️ Protein Digestion
☑️ Peptide Separation
☑️ Peptide Detection
☑️ Data Interpretation
The accompanying Peptide Coverage Map illustrates the binding site of AMG510 on the KRASG12C peptide. This map shows the peptide sequence LVVVGACGVGK, where the cysteine residue (Cys7) forms a covalent bond with AMG510. The fragment ions are color-coded based on their ion intensity, providing a detailed structural resolution of 1.0 residues.
☑️ Extensive Protein Library: Access a comprehensive protein library for rapid, customized assay development tailored to unique protein targets.
☑️ Assay Development and Optimization: Our experts optimize assays for high sensitivity and accuracy in detecting covalent binding interactions.
☑️ High Throughput Capacity: Process ~200 samples daily, ensuring fast, reliable results to meet your research timelines.
☑️ Advanced Instrumentation: Utilize cutting-edge tools like the Vanquish Flex LC system and QE Plus HRMS for precise, high-quality covalent binding data.
The GSH binding assay allows researchers to evaluate the formation of drug-GSH adducts and to better understand the covalent binding properties of potential therapeutic agents. By leveraging advanced mass spectrometry techniques, this assay provides detailed insights into the covalent interactions and stability of drug candidates, guiding the optimization of lead compounds for enhanced therapeutic efficacy. Afatinib, a covalent inhibitor targeting EGFR.
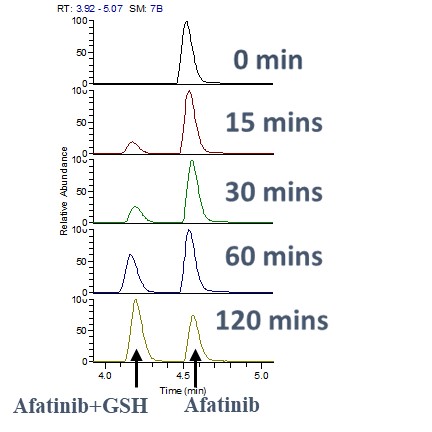
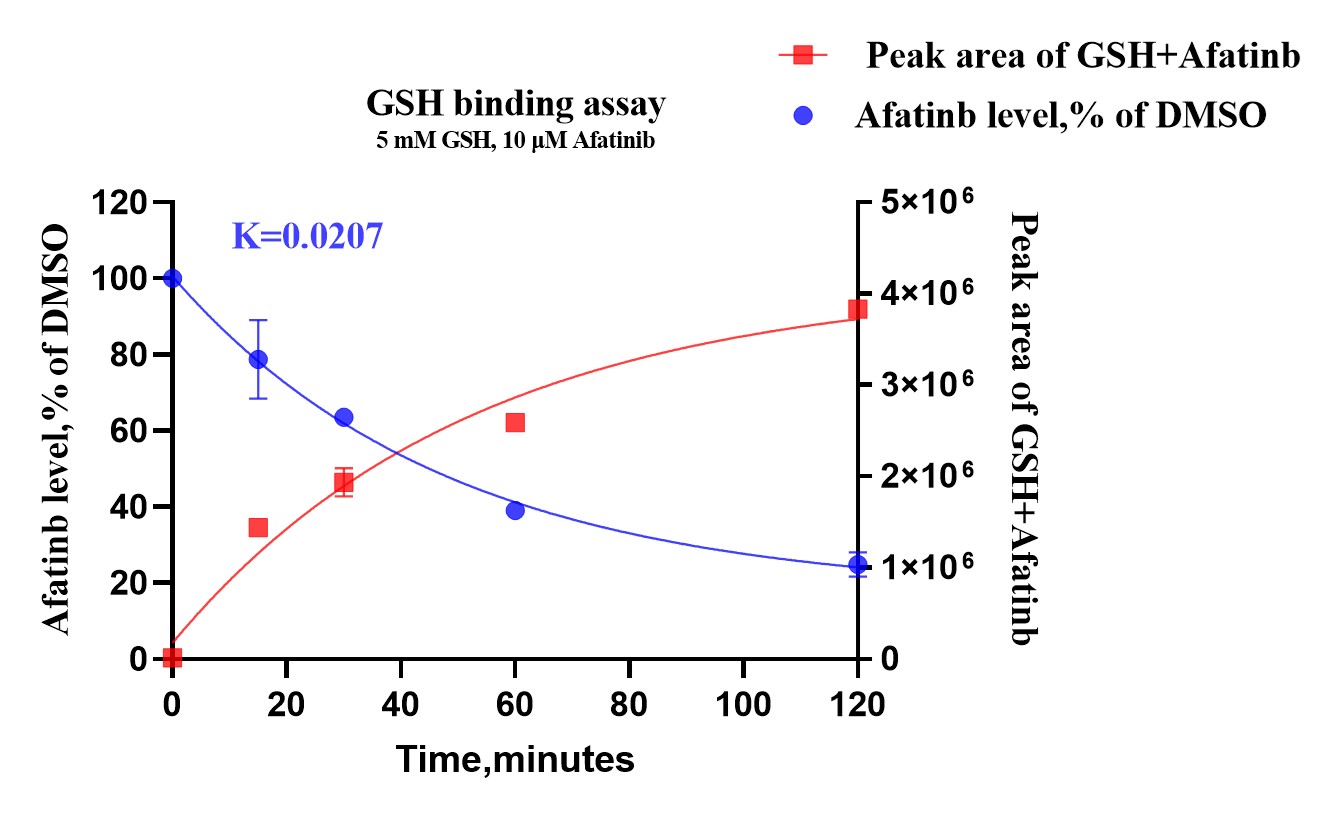
To determine the kinetics of the interaction, we monitored the reaction time between GSH and Afatinib. Samples were analyzed at various time intervals to observe the formation of Afatinib-GSH adducts. This assay helped in understanding the rate at which Afatinib forms covalent bonds with GSH, providing insights into its reactivity and stability.
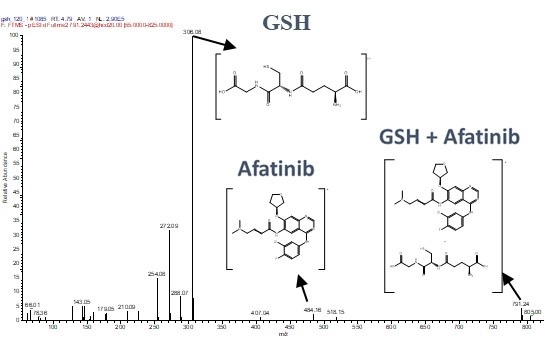
We utilized HRMS to analyze the fragmentation ion spectra of the Afatinib-GSH adduct. This detailed spectral analysis allowed us to identify and confirm the specific binding sites and the nature of the covalent bond formed. The high-resolution capabilities of the QE Plus HRMS provided precise molecular details, essential for understanding the binding mechanism at the atomic level.
To assess the stability and persistence of Afatinib in the presence of GSH, we measured the levels of Afatinib at different time points post-reaction. This assay enabled us to track the degradation or modification of Afatinib over time, ensuring a comprehensive understanding of its pharmacokinetic properties in a biological context.
We value your inquiries and are here to provide you with tailored solutions for your drug discovery and development needs. Whether you have questions, require more information, or are interested in discussing potential collaborations, our team of experts is just a message away.
Feel free to reach out to us.