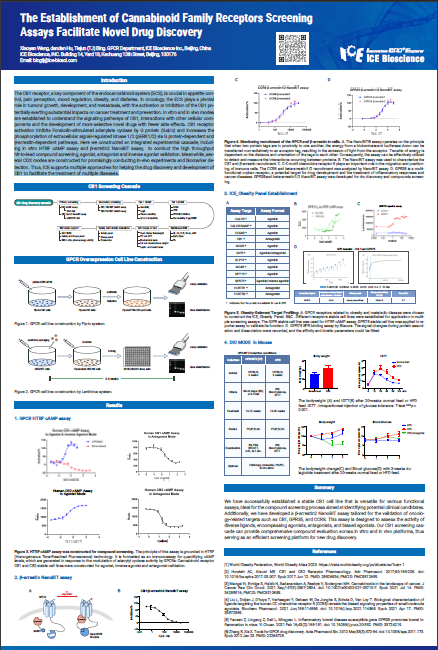
The CB1 receptor, a key component of the endocannabinoid system (ECS), is crucial in appetite control, pain perception, mood regulation, obesity, and diabetes. In oncology, the ECS plays a pivotal role in tumors' growth, development, and metastasis, with the activation or inhibition of the CB1 potentially exerting substantial impacts on cancer treatment and prevention. In vitro and in vivo modes are established to understand the signaling pathways of CB1, interactions with other cellular components and the development of more selective novel drugs with fewer side effects. CB1 receptor activation inhibits Forskolin-stimulated adenylate cyclase by G protein (Gαi/o) and increases the phosphorylation of extracellular signal-regulated kinase 1/2 (pERK1/2) via G protein-dependent and β-arrestin-dependent pathways. Here we constructed an integrated experimental cascade, including in vitro HTRF cAMP assay and β-arretin2 NanoBiT assay, to conduct the high throughput hit-to-lead compound screening, agonist, antagonist and inverse agonist validation. Meanwhile, several CDX modes are constructed for promisingly conducting in-vivo experiments and biomarker detection. Thus, ICE supports multiple approaches for helping the drug discovery and development of CB1 to facilitate the treatment of multiple diseases.
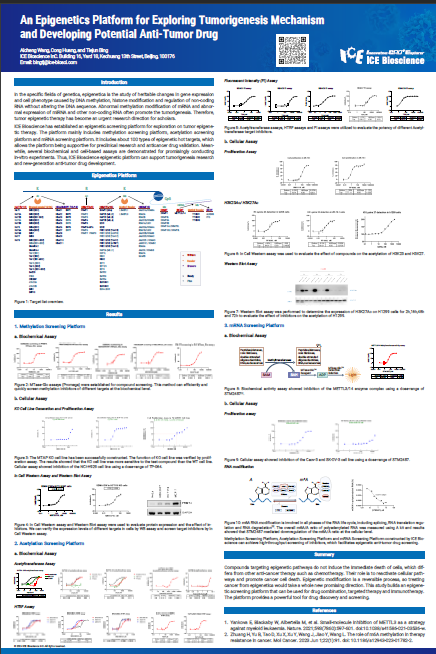
In the specific fields of genetics, epigenetics is the study of heritable changes in gene expression and cell phenotype caused by DNA methylation, histone modification and regulation of non-coding RNA without altering the DNA sequence. Abnormal methylation modification of mRNA and abnormal expression of miRNA and other non-coding RNA often promote the tumorigenesis. Therefore, tumor epigenetic therapy has become an urgent research direction for scholars.
ICE Bioscience has established an epigenetic screening platform for exploration on tumor epigenetic therapy. The platform mainly includes methylation screening platform, acetylation screening platform and mRNA screening platform. It includes about 100 types of epigenetic hot targets, which allows the platform being supportive for preclinical research and anticancer drug validation. Meanwhile, several biochemical and cell-based assays are demonstrated for promisingly conducting in-vitro experiments. Thus, ICE Bioscience epigenetic platform can support tumorigenesis research and new-generation anti-tumor drug development.

Antibody-drug conjugates (ADCs) are an innovative promising class of cancer therapeutics. ADCs are comprised three key components of an antibody (monoclonal antibody or bispecific antibody), a cytotoxic payload or other novel types of payload, and a linker. ADCs integrate chemotherapy and immunotherapy by combining the potency of payloads with the specificity of antibodies.
ICE Bioscience has established a biological and DMPK integrated platform for ADC screening and evaluation. The platform is dedicated to support a comprehensive service portfolio with various aspects of ADC development projects.
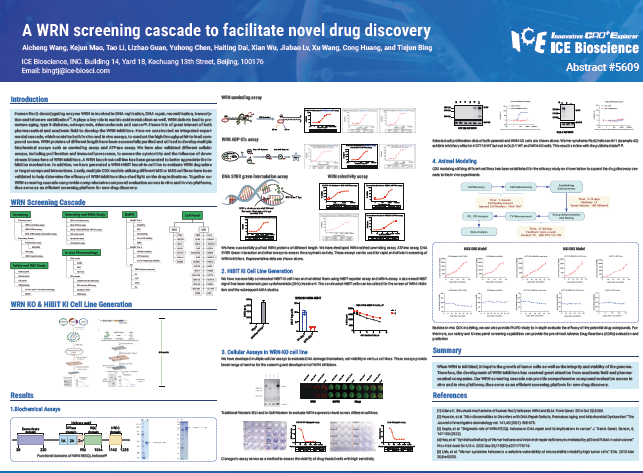
Human RecQ deconjugating enzyme WRN is involved in DNA replication, DNA repair, recombination, transcription and telomere stabilization. It plays a key role in nucleic acid metabolism as well. WRN defects lead to premature aging, type II diabetes, osteoporosis, atherosclerosis and cancer. Hence it is of great interest of both pharmaceutical and academic field to develop the WRN inhibitors. Here we constructed an integrated experimental cascade, which contains both in vitro and in vivo assays, to conduct the high throughput hit-to-lead compound screen. WRN proteins of different length have been successfully purified and utilized to develop multiple biochemical assays such as unwinding assay and ATPase assay. We have also validated different cellular assays, including proliferation and immunofluorescence, to assess the cytotoxicity and the influence of downstream biomarkers of WRN inhibitors. A WRN knock-out cell line has been generated to better appreciate the inhibition mechanism. In addition, we have generated a WRN-HiBiT knock-in cell line to evaluate WRN degraders or target-compound interactions. Lastly, multiple CDX models utilizing different MSI or MSS cell lines have been validated to help determine the efficacy of WRN inhibitors thus shed light on the drug indications. Together our WRN screening cascade can provide comprehensive compound evaluation across in vitro and in vivo platforms, thus serve as an efficient screening platform for new drug discovery.