Signal Transducer and Activator of Transcription 6 (STAT6) is a pivotal transcription factor within the JAK-STAT signaling pathway, activated downstream of cytokines IL-4 and IL-13. Upon phosphorylation at Tyr641, STAT6 undergoes reciprocal SH2 domain–mediated dimerization, translocates to the nucleus, and initiates transcription of genes central to Th2-type immune responses. Aberrant STAT6 signaling is implicated in asthma, atopic dermatitis, and other allergic diseases, making it a highly attractive target for therapeutic modulation.
While upstream blockade (e.g., via IL-4Rα antagonists) has shown clinical benefit, direct targeting of STAT6 is emerging as a promising alternative. Recent efforts have focused on a range of STAT6-binding molecules, including small-molecule inhibitors, PROTAC degraders, and peptide mimetics, many of which aim to disrupt the SH2 domain interactions critical for dimerization and function.
STAT6 consists of 847 amino acids and features a modular domain architecture:
1. N-terminal domain (ND, residues 1–130): Supports cooperative DNA binding and tetramerization
2. Coiled-coil domain (CCD): Mediates protein-protein interactions and nuclear import
3. DNA-binding domain (DBD): Recognizes GAS elements via a helix-turn-helix motif
4. Linker domain: Allows conformational flexibility
5. SH2 domain: Binds phosphorylated Tyr641 and mediates dimerization
6. Transactivation domain (TAD): Recruits coactivators (e.g., CBP/p300), subject to post-translational modifications
STAT6-IN-3: A small-molecule inhibitor blocking STAT6 phosphorylation or DNA binding for Th2-driven diseases.
AK-1690: A highly selective STAT6 PROTAC degrader with a DC50 of 1 nM, developed by the University of Michigan/Shaomeng Wang group.
KT-621: An oral STAT6 PROTAC degrader with picomolar potency against IL-4/IL-13 signaling, developed by Kymera Therapeutics for atopic dermatitis and asthma.
I-2: A reference compound used as a control in SPR assays.
To compare the binding site preferences of different STAT6-binding compounds, a SPR-based competitive binding assay was employed. The assay workflow is designed to distinguish whether two molecules bind to the same or distinct sites on STAT6:
Injection 1: Saturate the immobilized STAT6 protein with Compound A; monitor response unit (RU) until plateau
Injection 2: Inject a mixture of Compound A + Compound B
Note on Assay Design: In the second injection of the SPR competitive binding assay, a mixture of Compound A and Compound B is used—rather than Compound B alone—to ensure that the binding site occupied by Compound A remains saturated throughout the test. This setup allows accurate assessment of whether Compound B can bind to an independent site on the target protein. If Compound B binds a different site, the RU signal increases. If the two compounds compete for the same site, no additional binding is detected. This approach ensures reliable differentiation of overlapping vs. non-overlapping binding sites.
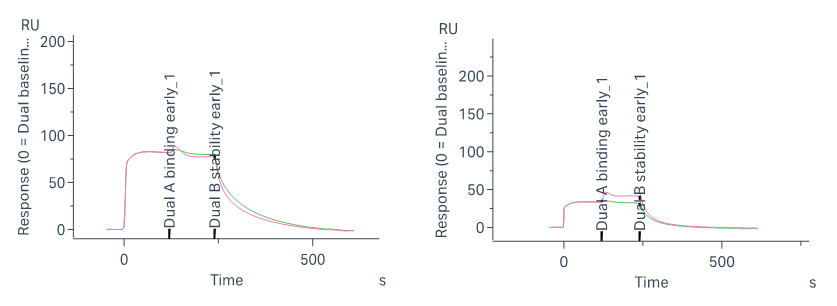
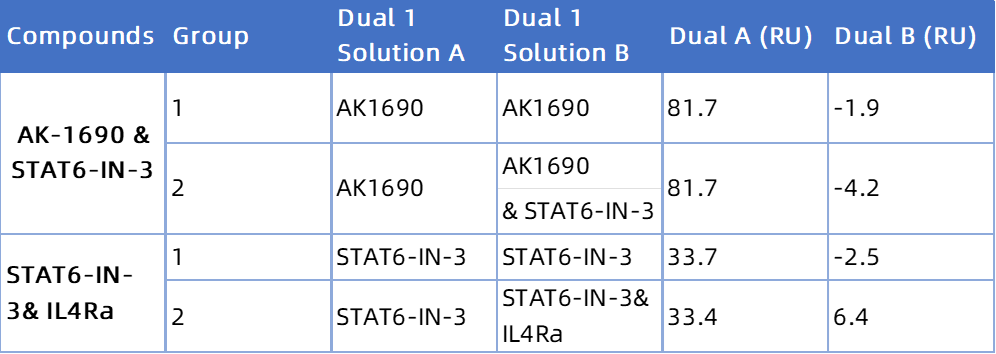
AK1690 & STAT6-IN-3: No RU increase → AK1690 and STAT6-IN-3 share the same binding site.
STAT6-IN-3 & IL-4Rα Peptide: RU increases → STAT6-IN-3 and IL-4Rα bind to distinct sites.
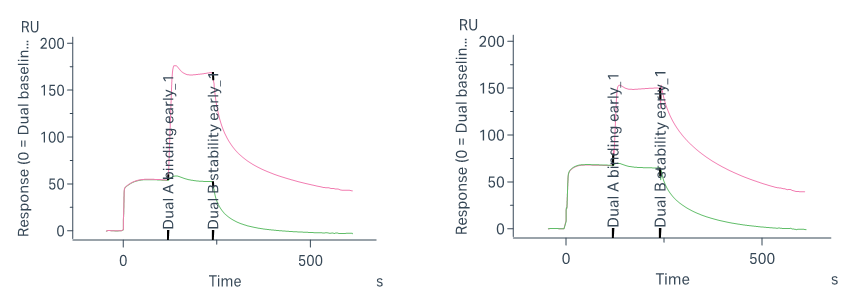
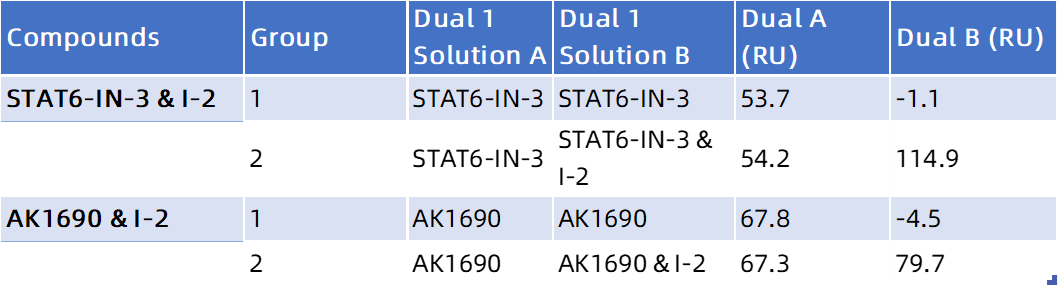
Significant RU increase → I-2 binds to a different site than both STAT6-IN-3 and AK1690.
This study demonstrates how SPR-based competition assays can accurately map binding site relationships among STAT6 inhibitors, even across different modalities (small molecules, PROTACs, peptides).
2025-10-30
2025-10-23
2025-09-28
2025-08-19
We value your inquiries and are here to provide you with tailored solutions for your drug discovery and development needs. Whether you have questions, require more information, or are interested in discussing potential collaborations, our team of experts is just a message away.
Feel free to reach out to us.