In our previous Application Note, ICE Bioscience highlighted the Amylin/CTR receptor axis, establishing CALCR and RAMP1/2/3 (AMY1–3) cell models and characterizing ligand pharmacology through HTRF cAMP assays with tool compounds such as hAmylin, hCT, Cagrilintide, and Petrelintide. This work provided a foundation for evaluating receptor activity and selectivity across the AMY/CTR family.
Building on that study, we now report two major advancements:
First, by benchmarking host cell responses, we identified COS7 as an optimal low-background system and established CTR and AMY1/2/3 lines to support high-throughput cAMP screening.
Second, we introduced β-arrestin recruitment assays using the NanoBiT system, generating HEK293T-Arrestin2-hCTR and -hAMY3 stable cells for quantitative arrestin signaling readouts.
Together, these extensions broaden ICE Bioscience’s GPCR toolkit, enabling deeper pharmacological insights and supporting drug discovery campaigns targeting the Amylin/CTR pathway.
To enable robust pharmacological profiling of the Amylin/Calcitonin receptor axis, ICE Bioscience systematically compared host cell responses to reference ligands including hCT, hAmylin, and the clinical analogue Cagrilintide using HTRF cAMP assays. While both HEK293 and Flp-In-CHO cells yielded obvious responses to hCT and Cagrilintide, COS7 cells remained unresponsive to either ligand. The absence of endogenous background activity in COS7 provided a clear baseline, making this cell line particularly suitable for engineering defined receptor models.
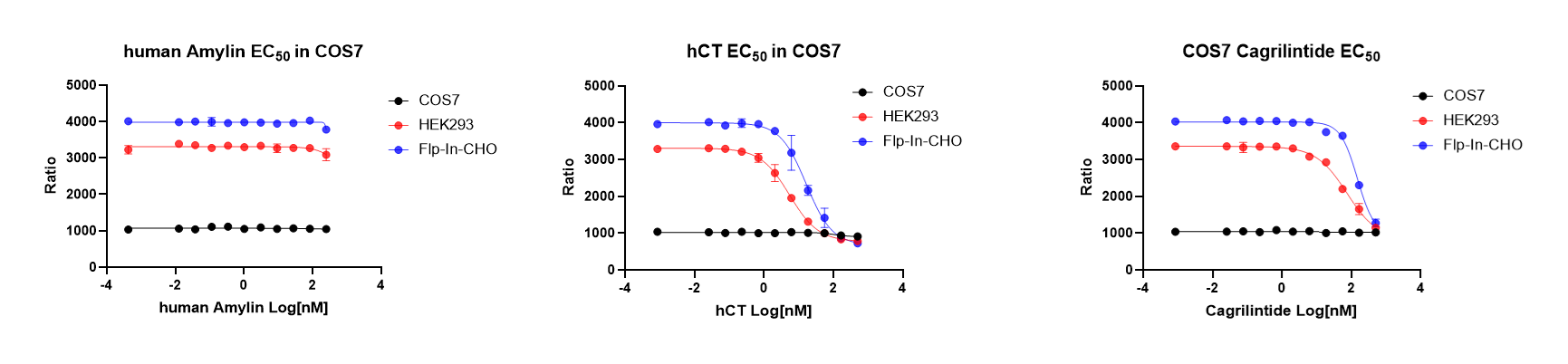
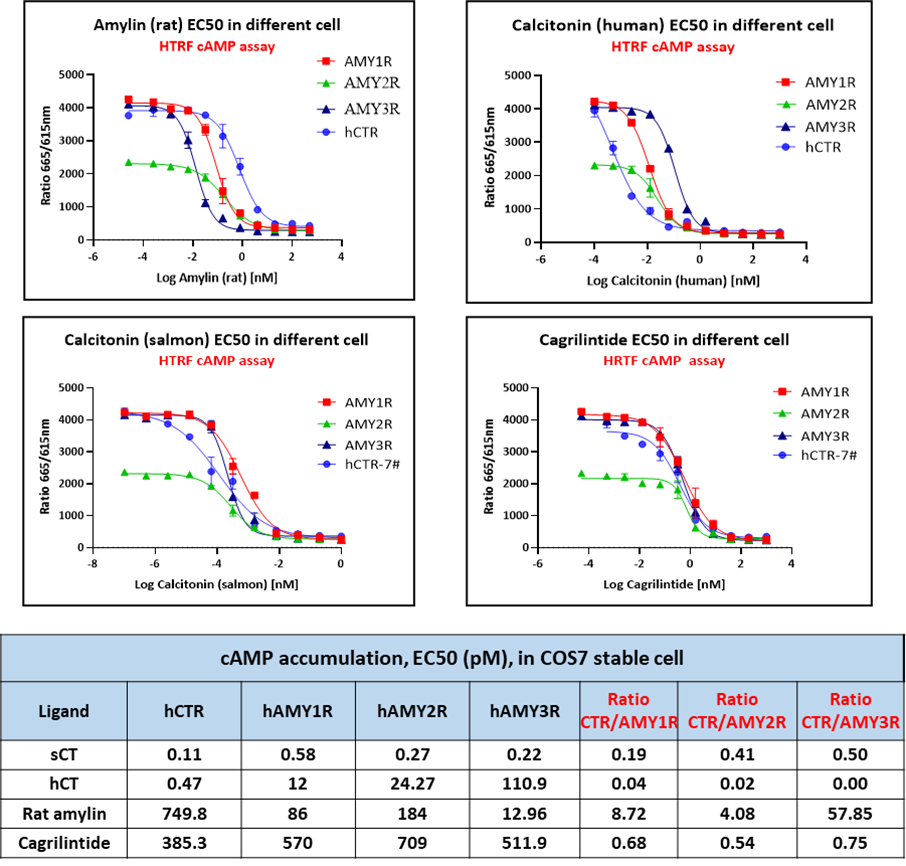
On this basis, ICE Bioscience established over-expressed COS7 monoclonal cell lines for CTR as well as AMY1, AMY2, and AMY3. These models offer a low-background, scalable system for high-throughput screening and pharmacological characterization.
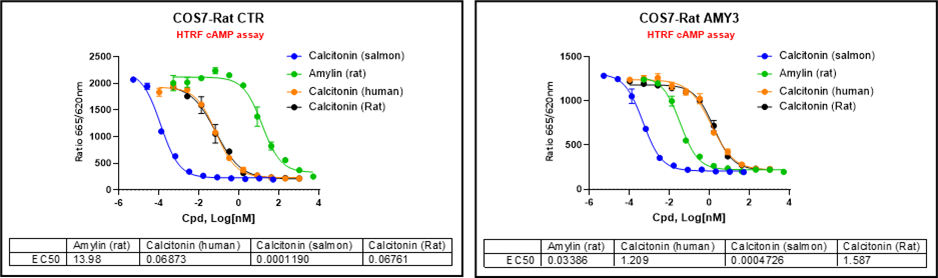
To further address species-selectivity requirements, COS7-based rat CTR and rat AMY3 monoclonal cell lines were generated. Using sCT, hCT, rat CT, and rat amylin as tool compounds, ICE Bioscience optimized HTRF cAMP assays that now provide a reliable, high-throughput platform for confirming compound activity at rat CTR and AMY3 in screening campaigns.
Upon agonist stimulation, the GPCR becomes phosphorylated at its C-terminal tail or intracellular loops, leading to the recruitment of β-arrestin 2 and the formation of a GPCR–β-arrestin 2 complex. In the NanoBiT system, the GPCR is fused to SmBiT (11 amino acids), and β-arrestin 2 is fused to LgBiT (17.6 kDa). Their proximity upon complex formation reconstitutes an active NanoLuc luciferase enzyme, generating a luminescent signal proportional to β-arrestin 2 recruitment. The β-arrestin 2 NanoBiT recruitment assay is a sensitive, quantitative, and high-throughput platform for assessing GPCR activation. It is broadly applicable in agonist/antagonist screening, biased signaling studies, and safety pharmacology evaluations.
ICE Bioscience's NanoBiT platform currently features 32 validated, screen-ready GPCR targets, supporting high-throughput drug discovery campaigns. Stable cell lines such as HEK293T-Arrestin2-hCTR and HEK293T-Arrestin2-hAMY3 have been developed and optimized for NanoBiT assays with ligands including sCT, hCT, and rat amylin.
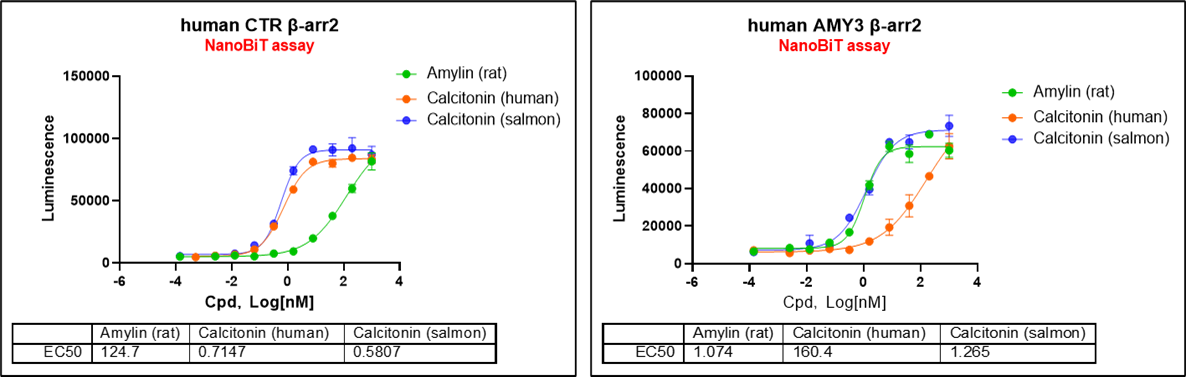
Together, these newly established COS7 and HEK293T-based cell models significantly expand ICE Bioscience’s GPCR assay capabilities across the Amylin/CTR receptor family. The integration of low-background cAMP assays and quantitative β-arrestin 2 recruitment readouts provides a dual-mode platform for dissecting ligand pharmacology, evaluating biased signaling, and supporting high-throughput screening in both human and rat receptor contexts.
The table below summarizes the currently available stable cell lines and assay formats, which are ready to support diverse research and drug discovery applications targeting the Amylin/CTR receptor axis.

2025-10-30
2025-10-23
2025-08-19
2025-08-15
We value your inquiries and are here to provide you with tailored solutions for your drug discovery and development needs. Whether you have questions, require more information, or are interested in discussing potential collaborations, our team of experts is just a message away.
Feel free to reach out to us.