Cyclin-dependent kinases (CDKs) play pivotal roles in regulating cell-cycle progression and transcriptional control, making them central targets in oncology and beyond. Traditional small-molecule inhibitors against CDKs have achieved notable clinical success, yet they often face limitations such as incomplete target suppression, acquired resistance, and off-target toxicities driven by conserved ATP-binding pockets.
In recent years, the emergence of targeted protein degradation has opened a new paradigm for modulating CDKs. Rather than simply blocking catalytic activity, CDK degraders—often designed as bifunctional molecules that recruit E3 ligases—can achieve complete elimination of disease-relevant kinase proteins, enabling sustained pathway silencing and the potential to overcome inhibitor-resistant mutations. This shift from inhibition to degradation fundamentally redefines how kinase biology can be explored and therapeutically exploited, creating new opportunities for precise, long-lasting modulation of the CDK network.
Accurate quantification of binary and ternary complex formation is essential for understanding the molecular basis of targeted protein degradation. In the discovery of CDK degraders,TR-FRET and spectral shift measurements provide rapid, sensitive, and quantitative means to evaluate the interaction between the degrader, the target kinase, and the recruited E3 ligase.
In this study, ICE Bioscience applied these complementary in vitro platforms to benchmark the formation efficiency of representative CDK degraders, including BSJ-04-132, a Ribociclib-based CDK4-selective Cereblon PROTAC; XY028-133, a VHL-recruiting CDK4/6 degrader; and CPS2, an irreversible, first-in-class CDK2 degrader.
TR-FRET Assays
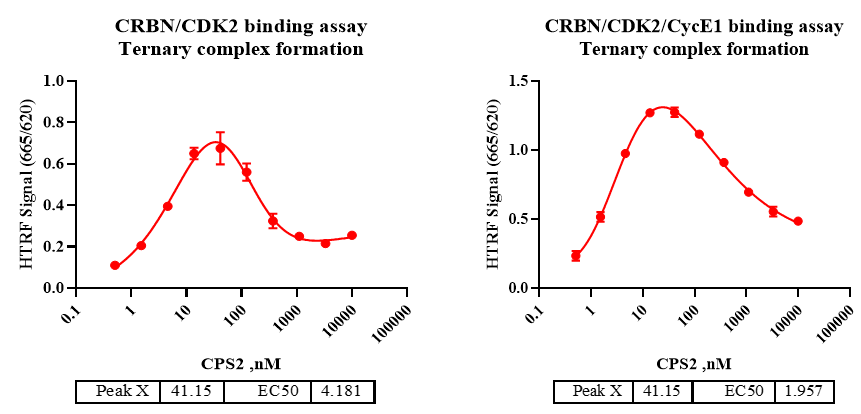
Inclusion of Cyclin E1 yields a higher maximal TR-FRET signal at a similar peak concentration but a lower EC50, indicating enhanced cooperativity/stability of the ternary complex when CDK2 is cyclin-bound.
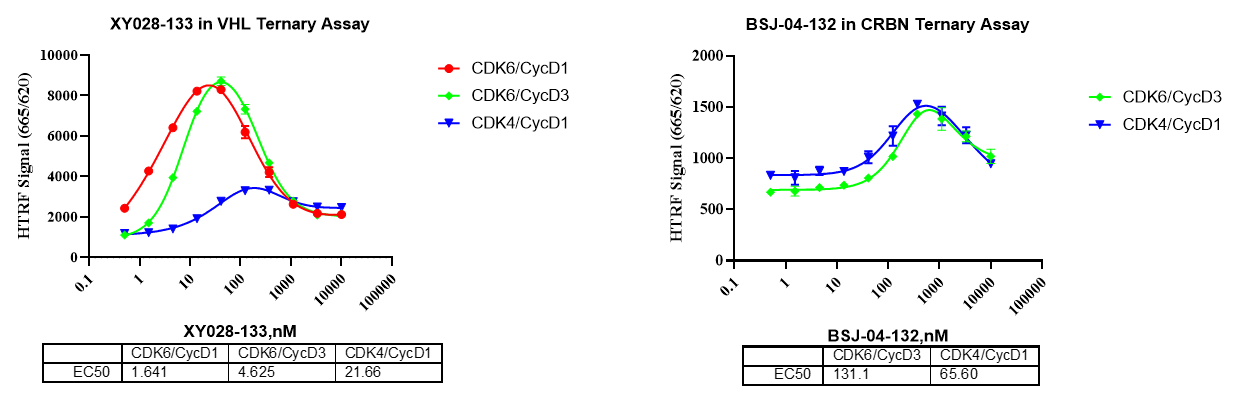
Together, these data demonstrate that distinct E3 ligase–recruiting scaffolds yield different ternary complex formation efficiencies.
Spectral Shift Assays
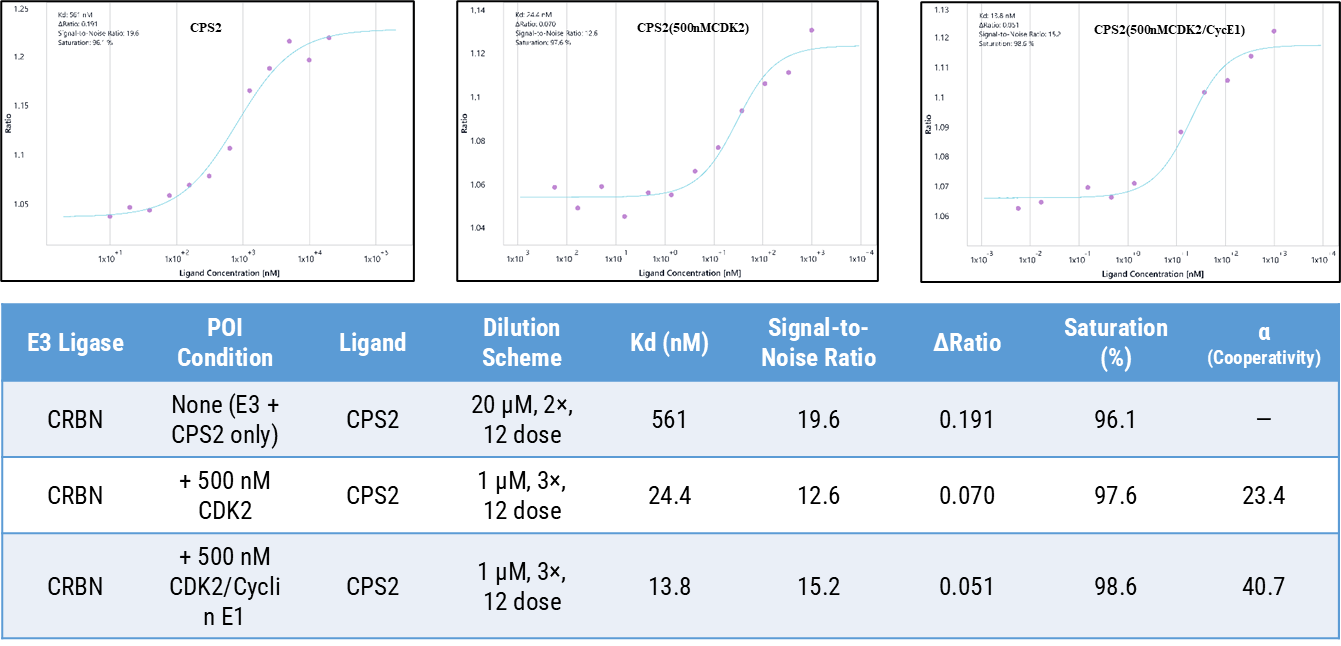
Spectral shift analysis quantitatively confirmed the cooperative interaction between CPS2, CRBN, and CDK2 in solution. In the absence of the POI, CPS2 bound CRBN with a modest affinity (Kd = 561 nM), consistent with the intrinsic binding strength of its cereblon-recruiting moiety. Upon addition of recombinant CDK2 (500 nM), the apparent affinity improved by more than twenty-fold (Kd = 24.4 nM; α = 23.4), indicating that engagement of CDK2 substantially enhances CPS2’s association with CRBN. When Cyclin E1 was introduced to form the CDK2/Cyclin E1 complex, affinity increased further (Kd = 13.8 nM; α = 40.7) together with a higher signal-to-noise ratio, demonstrating that the cyclin-bound conformation of CDK2 promotes a more stable and cooperative ternary configuration.
These results corroborate the TR-FRET findings: CPS2 preferentially stabilizes the ternary complex when CDK2 is in its active, Cyclin E1-associated state.
Collectively, the SPS and TR-FRET data establish that CPS2’s degradation efficiency is driven not only by its warhead-E3 affinity but also by structural cooperativity within the POI complex, underscoring the importance of conformational context in degrader optimization.

Western blot analysis was performed to confirm whether the cooperativity observed in vitro translates into target degradation in cells. CPS2 induced a clear, concentration-dependent reduction of CDK2 protein levels, while Vinculin remained constant as a loading control.
To extend the evaluation of CDK degraders beyond Western blot analysis, ICE Bioscience has established a HiBiT-tagged cellular assay platform enabling quantitative monitoring of target degradation. This system integrates stably expressed HiBiT fusion constructs for representative CDK family members such as CDK9 and Cyclin D1 (CCND1), allowing rapid and highly sensitive measurement of degradation kinetics in live cells.
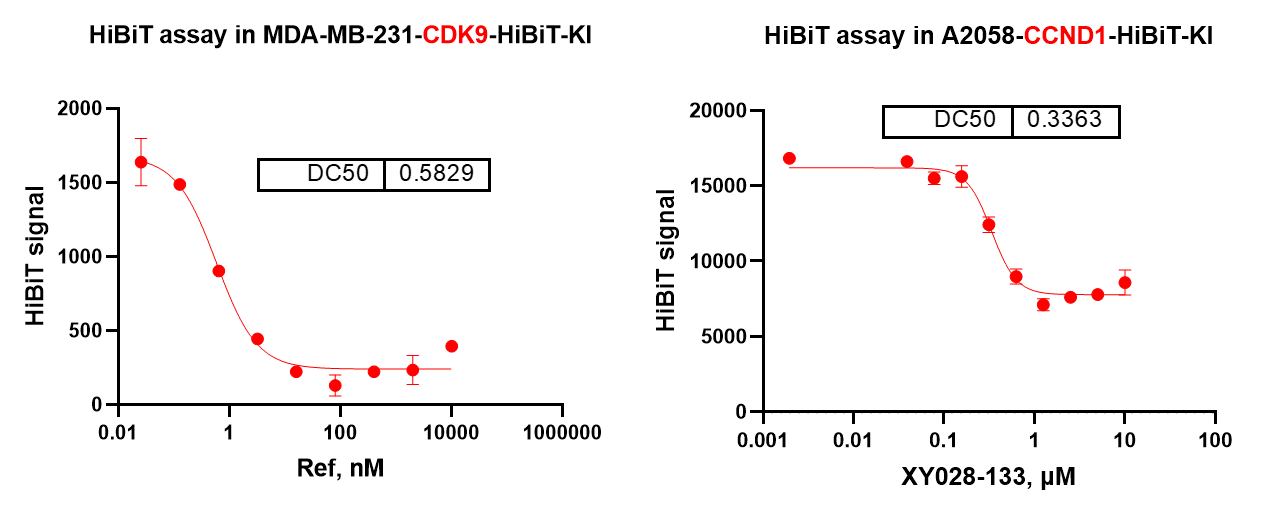
Using this luminescence-based readout, the platform provides accurate determination of DC₅₀ and Dmax, while supporting high-throughput compound profiling.
To ensure that targeted protein degraders achieve high specificity without triggering unintended substrate loss, ICE Bioscience has developed an integrated selectivity assessment platform that combines Western blot, HiBiT-based detection, and flow cytometry–based analysis. This multi-assay workflow enables quantitative evaluation of degrader selectivity across diverse cellular models and E3 ligase systems.
In summary, ICE Bioscience has established a workflow for the quantitative characterization of CDK-targeted degraders. By integrating biochemical and biophysical assays such as TR-FRET and spectral shift with cell-based validation including Western blot and HiBiT-based detection, the platform enables detailed evaluation of degrader-induced complex formation, cooperativity, and cellular potency. Complementary selectivity profiling assays further ensure that compound activity arises from specific E3-guided target degradation rather than non-selective substrate recruitment.
PREV:Integrated Profiling of STAT6 Degradatio...
NEXT:There's no more
2025-10-23
2025-09-28
2025-08-19
2025-08-15
We value your inquiries and are here to provide you with tailored solutions for your drug discovery and development needs. Whether you have questions, require more information, or are interested in discussing potential collaborations, our team of experts is just a message away.
Feel free to reach out to us.